5
Soft lithography is a new method of the micro-pattern replication process. e core is to
fabricate micro-structured chips through lithography and molding processes. It has the advantages
of low-cost and convenient processing, and is widely used in biotechnology, sensors, biotechnology,
and other elds. e processing steps of the mold are mainly as follows: rst, a layer of photoresist
is spin-coated on the hard substrates, then pre-baked to volatilize and solidify the solvent in the
photoresist, then subject to mask exposure and post-baking treatment, and nally a mold with
microstructure is developed.
With more chip materials discovered, the processing technologies of microuidic chips are
constantly updated. As the mainstream materials for processing microuidic chips, the molding
process of polymer materials will be further studied, and its application range will be broader.
1.2 SAMPLE MANIPULATION METHODS IN MICROFLUIDIC
CHIPS
e microuidic chip controls the ow of microuids through microuidic devices such as micro-
channels, microarrays, and microchambers; often combined with uidic methods [50, 51], magnetic
methods [52, 53], acoustic methods [54, 55 ], optical methods [56, 57], electrical methods [58, 59],
and other technical means to achieve micro-operation of biological samples.
1.2.1 FLUIDIC METHODS
Fluidic methods utilize special microstructures or microvalves in microuidic channels to control
microuidic ow to achieve manipulation of cells, such as single-cell capture [60, 61] and cell
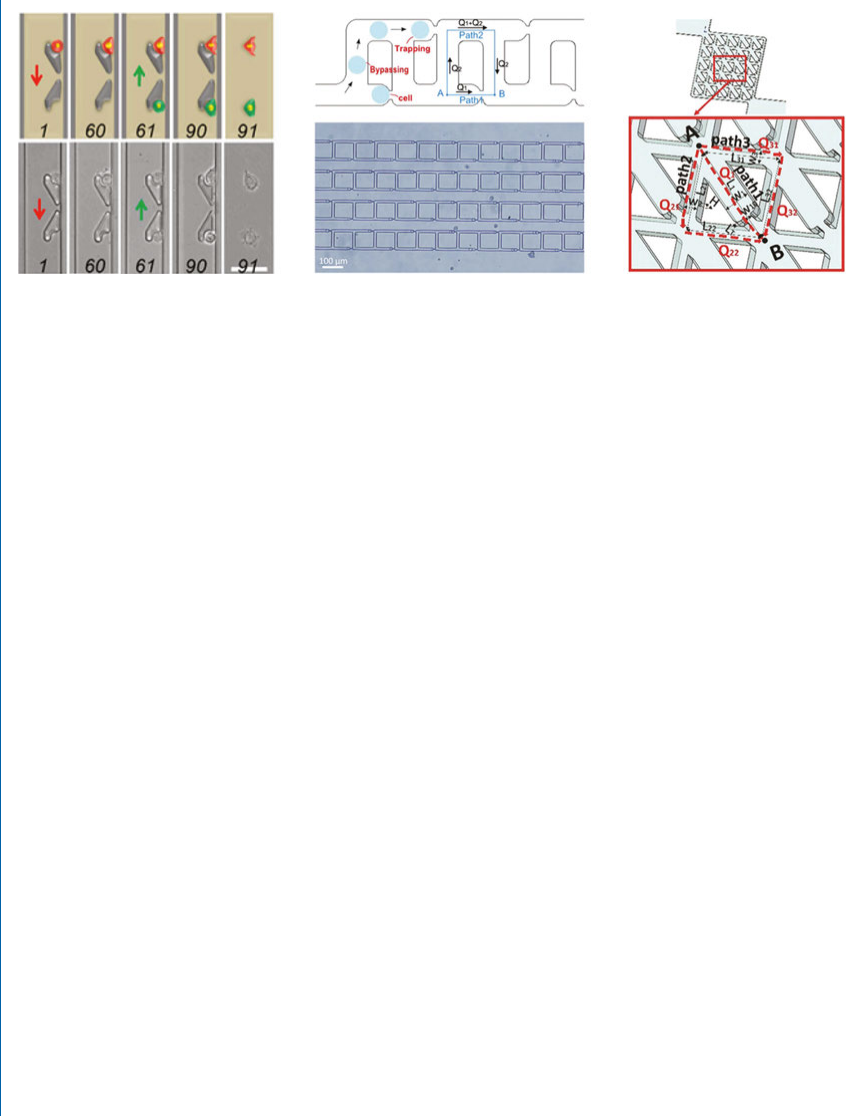
separation [62, 63]. For example, Zhang et al. designed a hook-type single-cell capture structure in
the microchannel, which uses a single-cell array capture structure to achieve capture and co-culture
of two cells (Figure 1.2(a)) [64]. In order to improve the eciency of single-cell capture, Jin et al.
designed a ladder-type single-cell capture chip based on the principle of minimum ow resistance,
with a single-cell capture eciency of 86% (Figure 1.2(b)) [65]. Also based on the principle of
minimum ow resistance, Mi et al. proposed a single-cell matrix capture structure (Figure 1.2(c)).
e device enables ecient deterministic single-cell capture and exible-cell capture patterning by
designing array patterns of trap site [66].
1.2 SAMPLE MANIPULATION METHODS IN MICROFLUIDIC CHIPS
6
1. INTRODUCTION
(a) (b) (c)
Figure 1.2: Single-cell capture structure: (a) hook-type capture structure [64]; (b) ladder-type capture
structure [65]; and (c) single-cell matrix capture structure [66]. Used with permission from the Royal
Society of Chemistry.
In cell separation, main methods include (a) inertial force [67, 68] and (b) deterministic lat-
eral displacement (DLD) [69, 70]. e inertial force methods use the characteristics of dierent size
cells in the spiral microchannel subject to dierent inertial forces and positional displacement to
achieve cell separation. Since the technology is based on a continuous ow, it is possible to process a
large number of samples in a short time. For example, Al-Halhouli et al. reported a label-free spiral
ow chip that focusing eciency of 99.1% of yeast cells (Figure 1.3(a)) [71]. e DLD methods are
to design micropillars array structures with a certain displacement angle in the microchannel, and
the continuous trajectory in the microchannel is dierent according to the dierence size to achieve
continuous separation. As cells ow along the streamline through the micropillars, larger cells
will shift laterally, and smaller cells will continue to ow along the original streamline. Compared
with the traditional membrane ltration methods, the DLD methods have the advantages of high
throughput, label-free separation, and no clogging. For example, Liu et al. proposed a microuidic
chip (Figure 1.3(b)) [72] that combines DLD array and surface antibody modication technology
to achieve rapid enrichment and capture of CTCs.
Although uid methods can realize label-free and high throughput biological manipulation,
the methods rely on the shape and structure of the microchannel, and depends on the dierences
of cell size that result in low specicity for cell sorting.
..................Content has been hidden....................
You can't read the all page of ebook, please click here login for view all page.
