2. An Overview of Biological Basics
A basic understanding of biology is essential to the biochemical engineer. Here we present selected aspects of basic biology at a level that will allow the student to function as a biochemical engineer. Although specific courses in biochemistry, genetics, microbiology, and cell biology provide much more depth, most engineering students do not have an opportunity to take three or four courses in the biological sciences, and some have no opportunity to take any of these courses. Thus, we have selected the parts of microbiology, biochemistry, genetics, and cell biology most directly relevant to bioprocesses to emphasize throughout chapters 2 to 8 and present this material with examples of application to bioprocesses.
2.1. Microbial Diversity
Life is very tenacious and can exist in extreme environments. Living cells can be found almost anywhere that water is in the liquid state. The right temperature, pH, and moisture levels vary from one organism to another.
Organisms from extreme environments (extremophiles) often provide the human race with important tools for processes to make useful chemicals and medicinals. They are also key to the maintenance of natural cycles and can be used in the recovery of metals from low-grade ores or in the desulfurization of coal or other fuels. The fact that organisms can develop the capacity to exist and multiply in almost any environment on earth is extremely useful.
Some cells can grow at –20°C (in a brine to prevent freezing), while others can grow at 120°C (where water is under high enough pressure to prevent boiling). Cells that grow best at low temperatures (below 20°C) are usually called psychrophiles, and those with temperature optima in the range of 20° to 50°C are mesophiles. Organisms that grow best at temperatures greater than 50°C are thermophiles.
Most organisms grow around neutral pH (6 to 8) and are called neutrophiles. There are some, however, that have pH optima far from neutrality, preferring pH values down to 1 or 2 (acidophiles) or well above pH 9 (alkaliphiles). Some organisms can grow at both low pH values and high temperatures. Although most organisms can grow only where water activity is high, others can grow on barely moist solid surfaces or in solutions with high salt concentrations (halophiles).
Some cells require oxygen for growth and metabolism. Such organisms can be termed aerobic. Other organisms are inhibited by the presence of oxygen and grow only anaerobically. Some organisms can switch the metabolic pathways to allow them to grow under either circumstance. Such organisms are facultative.
Often, organisms can grow in environments with almost no obvious source of nutrients. Some cyanobacteria (formerly called blue-green algae) can grow in an environment with only a little moisture and a few dissolved minerals. These bacteria are photosynthetic and can convert CO2 from the atmosphere into the organic compounds necessary for life. They can also convert N2 into NH3 for use in making the essential building blocks of life. Such organisms are important in colonizing nutrient-deficient environments.
Not only do organisms occupy a wide variety of habitats, but they also come in a wide range of sizes and shapes. Spherical, cylindrical, ellipsoidal, spiral, and pleomorphic cells exist. Special names are used to describe the shape of bacteria. A cell with a spherical or elliptical shape is often called a coccus (plural, cocci); a cylindrical cell is a rod or bacillus (plural, bacilli); a spiral-shaped cell is a spirillum (plural, spirilla). Some cells may change shape in response to changes in their local environment.
Thus, organisms can be found in the most extreme environments and have a wondrous array of different shapes, sizes, and metabolic capabilities. This great diversity provides the engineer with an immense variety of potential tools, many of which have been exploited. Take, for example, the heat-resistant enzyme Taq DNA polymerase, an enzyme which is derived from a thermophilic bacterium named Thermus aquaticus. Taq has become one of the most important enzymes in molecular biology because of its use in a DNA amplification technique known as the polymerase chain reaction (PCR; discussed in Chapter 8, “How Cellular Information Is Altered”).
2.1.1. Naming Cells
How cells are named is complicated by the bewildering variety of organisms present. A systematic approach to classifying these organisms is an essential aid to their intelligent use. Taxonomy is the development of approaches to organize and summarize our knowledge about the variety of organisms that exist. Although knowledge of taxonomy may seem remote from the needs of the engineer, it is necessary for efficient communication among engineers and scientists working with living cells. Taxonomy can also play a critical role in patent litigation involving bioprocesses.
While taxonomy is concerned with approaches to classification, nomenclature refers to the actual naming of organisms. For microorganisms, we use a dual name (binary nomenclature). The names are given in Latin, or are Latinized. A genus is a group of related species, and a species includes organisms that are substantially alike. A common gut organism that has been well studied is Escherichia coli. Escherichia is the genus and coli the species. When writing a report or paper, it is common practice to give the full name when the organism is first mentioned but in subsequent discussion to abbreviate the genus to the first letter. In this case, we would use E. coli. Although organisms that belong to the same species all share the same major characteristics, there are subtle and often technologically important variations within species. An E. coli used in one laboratory may differ from that used in another. Thus, various strains and substrains are designated by the addition of letters and numbers. For example, E. coli B/r A will differ in growth and physiological properties from E. coli K12.
Now that we know how to name organisms, we could consider broader classification up to the level of kingdoms. There is no universal agreement on how to classify microorganisms at this level. Such classification is rather arbitrary and need not concern us. However, we must be aware that there are two primary cell types: eucaryotic and procaryotic. The primary difference between them is the presence or absence of a membrane around the cell’s genetic information.
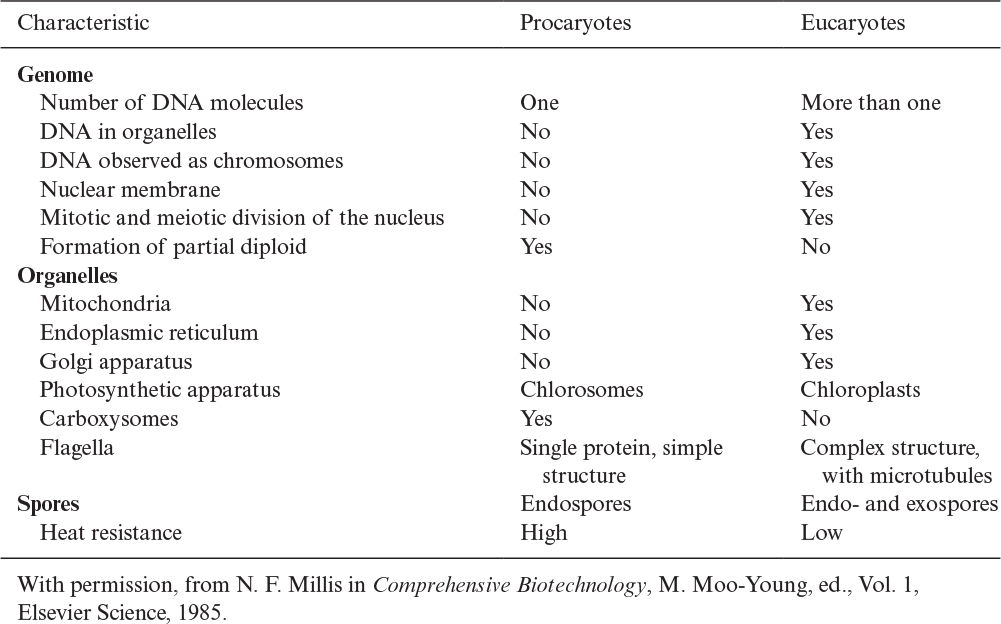
Procaryotes have a simple structure with a single chromosome. Procaryotic cells have no nuclear membrane and no organelles, such as the mitochondria and endoplasmic reticulum. Eucaryotes have a more complex internal structure, with more than one chromosome (DNA molecule) in the nucleus. Eucaryotic cells have a true nuclear membrane and contain mitochondria, endoplasmic reticulum, Golgi apparatus, and a variety of specialized organelles. We describe each of these components in Section 2.1.4. A detailed comparison of procaryotes and eucaryotes is presented in Table 2.1. Structural differences between procaryotes and eucaryotes are discussed later.
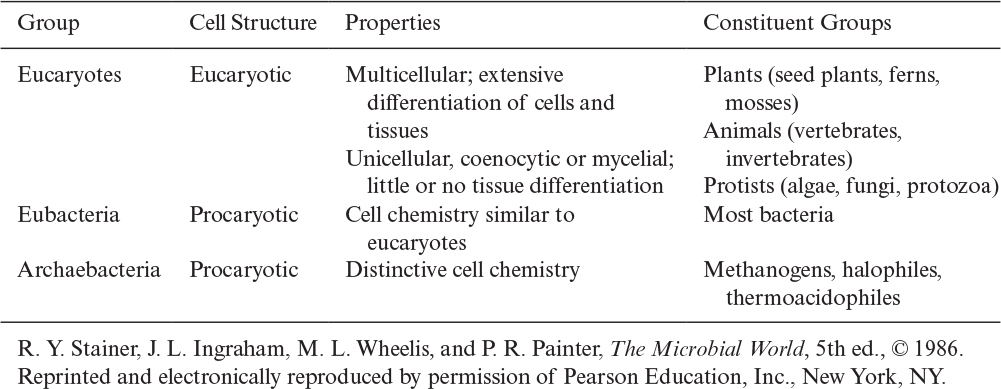
There is further distinction of subgroups within procaryotes. Evidence suggests that a common or universal ancestor gave rise to three distinctive branches of life: eucaryotes, eubacteria (or “true” bacteria), and archaebacteria. Table 2.2 summarizes some of the distinctive features of these groups. All of the genes of whole organisms, known as the genome, can now be readily sequenced and this information is having a great impact on our understanding of how these families evolved and are related.
Viruses cannot be classified under any of these categories, as they are not free-living organisms. Let’s consider first some of the characteristics of these rather simple “organisms.”
2.1.2. Viruses
Viruses are very small and are obligate parasites of other cells, such as bacterial, yeast, plant, and animal cells. Viruses cannot capture or store free energy and are not functionally active except when inside their host cells. The sizes of viruses vary from 30 to 200 nanometers (nm). Viruses contain either DNA (DNA viruses) or RNA (RNA viruses) as genetic material. DNA stands for deoxyribonucleic acid, and RNA is ribonucleic acid; we will soon discuss these molecules in more detail. In free-living cells, all genetic information is contained in the DNA, whereas viruses can use either RNA or DNA to encode such information. The viral genome is surrounded by a protein coat called a capsid. Some viruses have an outer envelope of a lipoprotein and some do not.
Almost all cell types are susceptible to viral infections. Viruses infecting bacteria are called bacteriophages. Some bacteriophages have a hexagonal head, tail, and tail fibers. Bacteriophages attach to the cell wall of a host cell with tail fibers, alter the cell wall of the host cell, and inject the viral genetic material into the host cell. Figure 2.1 describes the attachment of a virus onto a host cell. Bacteriophage nucleic acids reproduce inside the host cells to produce more phages. At a certain stage of viral reproduction, host cells lyse, or break apart, and phage particles are released, which can infect new host cells. This mode of reproduction of viruses is called the lytic cycle. In some cases, phage DNA may be incorporated into the host DNA, and the host may continue to multiply in this state, which is called the lysogenic cycle.

Figure 2.1. Replication of a virulent bacteriophage. A virulent phage undergoes a lytic cycle to produce new phage particles within a bacterial cell. Cell lysis releases new phage particles that can infect more bacteria. (J. G. Black, Microbiology: Principles and Applications, 3d ed., © 1996.)
Viruses are the cause of many diseases, and antiviral agents are important targets for drug discovery. Additionally, viruses are directly important to bioprocess technology. For example, a phage attack on an E. coli fermentation to make a recombinant protein product can be extremely destructive, causing the loss of the entire culture in vessels of many thousands of liters. However, phages can be used as agents to move desired genetic material into E. coli. Modified animal viruses can be used as vectors in genetically engineering animal cells to produce proteins from recombinant DNA technology. In some cases, a killed virus preparation is used as a vaccine. In other cases, genetic engineering allows production of viruslike particles (VLPs) that are empty shells; the shell is the capsid, and all nucleic acid is removed. Such particles can be used as vaccines without fear of viral replication because all of the genetic material has been removed. For gene therapy, one approach is to use a virus in which viral genetic material has been modified or replaced with the desired gene to be inserted into the patient. The viral capsid can act as a Trojan horse to protect the desired gene in a hostile environment and then deliver it selectively to a particular cell type. Thus, although viruses can do great harm, they are also important biotechnological tools.
2.1.3. Procaryotes
The sizes of most procaryotes vary from 0.5 to 3 micrometers (μm) in equivalent radius. Different species have different shapes, such as spherical or coccus (e.g., Staphylococci), cylindrical or bacillus (e.g., E. coli), or spiral or spirillum (e.g., Rhodospirillum). Procaryotic cells grow rapidly, with typical doubling times of less than 30 minutes to several hours. Also, procaryotes can utilize a variety of nutrients as carbon source, including carbohydrates, hydrocarbons, proteins, and CO2.
2.1.3.1. Eubacteria.
Eubacteria can be divided into several different groups. One distinction is based on the Gram stain (developed by Hans Christian Gram in 1884). The staining procedure first requires fixing the cells by heating. The basic dye, crystal violet, is added; all bacteria will stain purple. Next, iodine is added, followed by the addition of ethanol. Gram-positive cells remain purple, whereas gram-negative cells become colorless. Finally, counterstaining with safranin leaves gram-positive cells purple, whereas gram-negative cells turn red. The ability to react with the Gram stain reveals intrinsic differences in the structure of the cell envelope.
A typical gram-negative cell is E. coli (see Figure 2.2). It has an outer membrane supported by a thin peptidoglycan layer. Peptidoglycan is a complex polysaccharide with amino acids and forms a structure somewhat analogous to a chain-link fence. A second membrane (the inner or cytoplasmic membrane) exists and is separated from the outer membrane by the periplasmic space. The cytoplasmic membrane contains by weight about 50% protein, 30% lipids, and 20% carbohydrates. The cell envelope serves to retain important cellular compounds and to preferentially exclude undesirable compounds in the environment. Lipopolysaccharide (LPS), also known as lipoglycans and endotoxins, are macromolecules consisting of a lipid covalently linked to a polysaccharide composed of O-antigen, outer core, and inner core; they are found in the outer membrane of gram-negative bacteria and elicit strong immune responses in animals. Consequently, LPS removal is a key consideration when using gram-negative bacteria such as E. coli for producing therapeutic molecules. Conversely, LPS and its subcomponents (e.g., O-antigen) are the focus of numerous vaccine development efforts. Loss of membrane integrity leads to cell lysis (cells breaking open) and cell death. The cell envelope is crucial to the transport of selected material in and out of the cell.
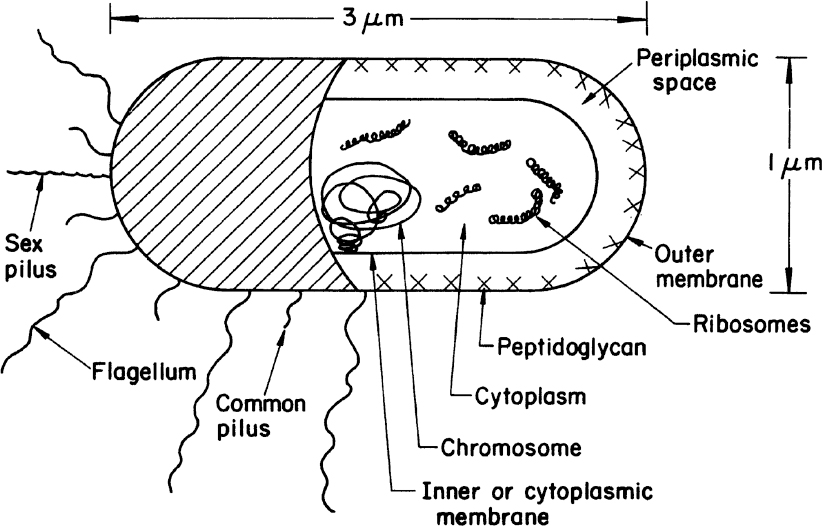
Figure 2.2. Schematic of a typical gram-negative bacterium (E. coli). A gram-positive cell would be similar, except that it would have no outer membrane, its peptidoglycan layer would be thicker, and the chemical composition of the cell wall would differ significantly from the outer envelope of the gram-negative cell.
A typical gram-positive cell is Bacillus subtilis. Gram-positive cells do not have an outer membrane. Rather, they have a very thick, rigid cell wall with multiple layers of peptidoglycan. Gram-positive cells also contain teichoic acids covalently bonded to the peptidoglycan. Because gram-positive bacteria have only a cytoplasmic membrane, they are often much better suited to excretion of proteins into the culture media. Such excretion can be technologically advantageous when the protein is a desired product.
Some bacteria are not gram-positive or gram-negative. For example, the Mycoplasma genus of bacteria have no cell walls. These bacteria are important not only clinically (e.g., primary atypical pneumonia) but also because they commonly contaminate media used industrially for animal cell culture.
Actinomycetes are bacteria, but, morphologically, they resemble molds with their long and highly branched hyphae. However, the lack of a nuclear membrane and the composition of the cell wall require classification as bacteria. Actinomycetes are important sources of antibiotics. Some possess amylolytic and cellulolytic enzymes and are effective in enzymatic hydrolysis of starch and cellulose. Actinomyces, Thermomonospora, and Streptomyces are examples of genera belonging to this group.
Other distinctions within the eubacteria can be made on the basis of cellular nutrition and energy metabolism. One important example is photosynthesis. The cyanobacteria (formerly called blue-green algae) have chlorophyll and fix CO2 into sugars. Anoxygenic photosynthetic bacteria (the purple and green bacteria) have light-gathering pigments called bacteriochlorophyll. Unlike true photosynthesis, the purple and green bacteria do not obtain reducing power from the splitting of water and do not form oxygen.
When stained properly, the area occupied by the procaryotic cell’s DNA can be easily seen. Procaryotes may also have other visible structures when viewed under the microscope, such as ribosomes, storage granules, bacterial microcompartments (BMCs), spores, and volutins. Ribosomes are the site of protein synthesis. A typical bacterial cell contains approximately 10,000 ribosomes per cell, although this number can vary greatly with growth rate. A ribosome typically is 10 to 20 nm in size and consists of approximately 63% RNA and 37% protein.
Storage granules (which are not present in every bacterium) can be used as a source of key metabolites and often contain polysaccharides, lipids, phosphate, and sulfur granules. The sizes of storage granules vary between 0.5 and 1 μm. Along similar lines, BMCs are organelles that enclose enzymes and other proteins. BMCs are typically about 40 to 200 nm in diameter and are made entirely of proteins that self-assemble into icosahedral or quasi-icosahedral structures. The protein shell of BMCs is selectively permeable, functioning like a membrane. The majority are involved in either carbon fixation (carboxysomes) or aldehyde oxidation (metabolosomes), and efforts to engineer them to contain customized cargo are currently being pursued.
Some bacteria make intracellular spores (often called endospores in bacteria). Bacterial spores are produced as a resistance to adverse conditions such as high temperature, radiation, and toxic chemicals. The usual concentration is 1 spore per cell, with a spore size of about 1 μm. Spores can germinate under favorable growth conditions to yield actively growing bacteria.
Volutin is another granular intracellular structure, made of inorganic polymetaphosphates, that is present in some species. Some photosynthetic bacteria, such as Rhodospirillum, have chromatophores that are large inclusion bodies (50 to 100 nm) utilized in photosynthesis for the absorption of light.
Extracellular products can adhere to or be incorporated within the surface of the cell. Certain bacteria have a coating or outside cell wall called capsule, which is usually a polysaccharide or sometimes a polypeptide. Extracellular polymers are important to biofilm formation and response to environmental challenges (e.g., viruses). Similar to LPS, capsular polysaccharides of pathogenic bacteria have also become an important target for vaccine development. Table 2.3 summarizes the architecture of most bacteria.
2.1.3.2. Archaebacteria.
The archaebacteria appear under the microscope to be nearly identical to many of the eubacteria. However, these cells differ greatly at the molecular level. In many ways, the archaebacteria are as similar to the eucaryotes as they are to the eubacteria. Some examples of differences between archaebacteria and eubacteria are as follows:
• Archaebacteria have no peptidoglycan.
• The nucleotide sequences in the ribosomal RNA are similar within the archaebacteria but distinctly different from eubacteria.
• The lipid composition of the cytoplasmic membrane is very different for the two groups.
The archaebacteria usually live in extreme environments and possess unusual metabolism. Methanogens, which are methane-producing bacteria, as well as the thermoacidophiles and halobacteria, belong to this group. The thermoacidophiles can grow at high temperatures and low pH values, whereas the halobacteria can live only in very strong salt solutions. Together, these organisms are important sources for catalytically active proteins (enzymes) with novel properties.
2.1.4. Eucaryotes
Fungi (yeasts and molds), algae, protozoa, and animal and plant cells constitute the eucaryotes. Eucaryotes are five to ten times larger than procaryotes in diameter (e.g., yeast about 5 μm, animal cells about 10 μm, and plants about 20 μm). Eucaryotes have a true nucleus and a number of cellular organelles inside the cytoplasm. Figure 2.3 is a schematic of two typical eucaryotic cells.
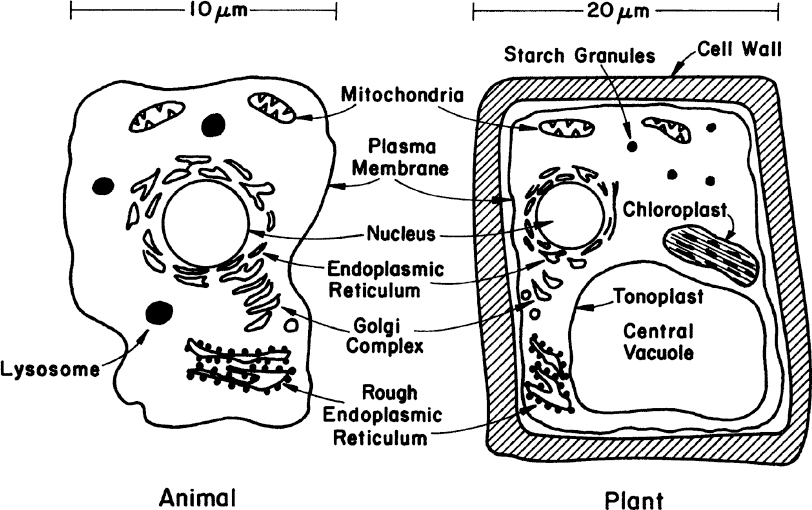
Figure 2.3. The two primary types of higher eucaryotic cells. Neither sketch is complete, but each summarizes the principal differences and similarities of such cells.
In cell-wall and cell-membrane structure, eucaryotes are similar to procaryotes. The plasma membrane is made of proteins and phospholipids that form a bilayer structure. Major proteins of the membrane are hydrophobic and are embedded in the phospholipid matrix. One major difference is the presence of sterols in the cytoplasmic membrane of eucaryotes. Sterols strengthen the structure and make the membrane less flexible. The cell wall of eucaryotic cells shows considerable variations. Some eucaryotes have a peptidoglycan layer in their cell wall; some have polysaccharides and cellulose (e.g., algae). The plant cell wall is composed of cellulose fibers embedded in pectin aggregates, which impart strength to the cell wall. Animal cells do not have a cell wall but only a cytoplasmic membrane. For this reason, animal cells are very shear-sensitive and fragile. This factor significantly complicates the design of large-scale bioreactors for animal cells.
The nucleus of eucaryotic cells contains chromosomes as nuclear material (DNA molecules with some closely associated small proteins), surrounded by a membrane. The nuclear membrane consists of a pair of concentric and porous membranes. The nucleolus is an area in the nucleus that stains differently and is the site of ribosome synthesis. However, many chromosomes contain small amounts of RNA and basic proteins called histones attached to the DNA. Each chromosome contains a single linear DNA molecule on which the histones are attached.
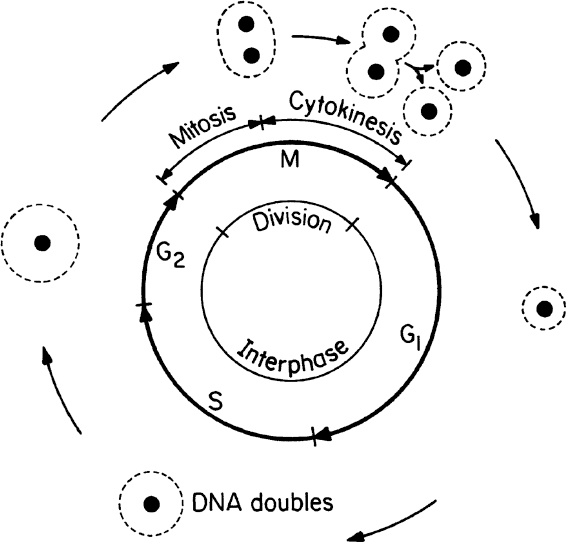
Cell division (asexual) in eucaryotes involves several major steps, such as DNA synthesis, nuclear division, cell division, and cell separation. Sexual reproduction in eucaryotic cells involves the conjugation of two cells called gametes (egg and sperm cells). The single cell formed from the conjugation of gametes is called a zygote. The zygote has twice as many chromosomes as does the gamete. Gametes are haploid cells, while zygotes are diploid. For humans, a haploid cell contains 23 chromosomes, and diploid cells have 46. The cell-division cycle (asexual reproduction) in a eucaryotic cell is depicted in Figure 2.4.
The cell-division cycle is divided into four phases. The M phase consists of mitosis whereby the nucleus divides and cytokinesis whereby the cell splits into separate daughter cells. All of the phases between one M phase and the next are known collectively as the interphase. The interphase is divided into three phases: G1, S, and G2. The cell increases in size during the interphase period. In the S phase, the cell replicates its nuclear DNA. There are key checkpoints in the cycle when the cell machinery must commit to entry into the next phase. Checkpoints exist for entry into the S and M phases and exit from M phase. Cells may also be in a G0 state, which is a resting state in which no growth occurs.
The mitochondria are the powerhouses of a eucaryotic cell, where respiration and oxidative phosphorylation take place. Mitochondria have a nearly cylindrical shape 1 μm in diameter and 2 to 3 μm in length. The typical structure of a mitochondrion is shown in Figure 2.5. The external membrane is made of a phospholipid bilayer with proteins embedded in the lipid matrix. The mitochondria contain a complex system of inner membranes called cristae. A gel-like matrix containing large amounts of protein fills the space inside the cristae. Some enzymes of oxidative respiration are bound to the cristae. A mitochondrion has its own DNA and protein-synthesizing machinery and reproduces independently.

Figure 2.5. The structure of mitochondria as seen in electron micrographs of thin sections of eucaryotic cells. (R. Y. Stainer, J. L. Ingraham, M. L. Wheelis, and P. R. Painter, The Microbial World, 5th ed., © 1986. Reprinted and electronically reproduced by permission of Pearson Education, Inc., New York, NY.)
The endoplasmic reticulum is a complex, convoluted membrane system leading from the cell membrane into the cell. The rough endoplasmic reticulum contains ribosomes on the inner surfaces and is the site of protein synthesis and modifications of protein structure after synthesis. The smooth endoplasmic reticulum is more involved with lipid synthesis.
Lysosomes are very small, membrane-bound particles that contain and release digestive enzymes. Lysosomes contribute to the digestion of nutrients and invading substances.
Peroxisomes are similar to lysosomes in their structure but not in function. Peroxisomes carry out oxidative reactions that produce hydrogen peroxide.
Glyoxysomes are also very small, membrane-bound particles that contain the enzymes of the glyoxylate cycle.
Golgi bodies are very small particles composed of membrane aggregates and are responsible for the secretion of certain proteins. Golgi bodies are sites where proteins are modified by the addition of various complex sugars known as glycans in a process called glycosylation. Such modifications are important to protein function in the body.
Vacuoles are membrane-bound organelles of low density and are responsible for food digestion, osmotic regulation, and waste-product storage. Vacuoles may occupy a large fraction of cell volume (up to 90% in plant cells).
Chloroplasts are relatively large, chlorophyll-containing, green organelles that are responsible for photosynthesis in photosynthetic eucaryotes, such as algae and plant cells. Every chloroplast contains an outer membrane and a large number of inner membranes called thylakoids. Chlorophyll molecules are associated with thylakoids, which have a regular membrane structure with lipid bilayers. Chloroplasts are autonomous units containing their own DNA and protein-synthesizing machinery.
Certain procaryotic and eucaryotic organisms contain flagella—long, filamentous structures that are attached to one end of the cell and are responsible for the motion of the cell. Eucaryotic flagella contain 2 central fibers surrounded by 18 peripheral fibers, which exist in doublets. Fibers are in a tube structure called a microtubule and are composed of proteins called tubulin. The whole fiber assembly is embedded in an organic matrix and is surrounded by a membrane.
The cytoskeleton (in eucaryotic cells) consists of filaments that provide an internal framework to organize the cell’s internal activities and control its shape. These filaments are critical in cell movement, transduction of mechanical forces into biological responses, and separation of chromosomes into the two daughter cells during cell division. Three types of fibers are present: actin filaments, intermediate filaments, and microtubules.
Cilia are flagella-like structures but are numerous and shorter. Only one group of protozoa, called ciliates, contains cilia. Paramecium species contain nearly 104 cilia per cell. Ciliated organisms move much faster than flagellated ones.
This completes our summary of eucaryotic cell structure. Now let us turn our attention to the microscopic eucaryotes.
Fungi are heterotrophs that are widespread in nature. Fungal cells are larger than bacterial cells, and their typical internal structures, such as nucleus and vacuoles, can be seen easily with a light microscope. Two major groups of fungi are yeasts and molds.
Yeasts are single small cells of 5 to 10 μm in size. Yeast cells are usually spherical, cylindrical, or oval. Yeasts can reproduce by asexual or sexual means. Asexual reproduction is by either budding or fission. In budding, a small bud cell forms on the cell and gradually enlarges and separates from the mother cell. Asexual reproduction by fission is similar to that of bacteria. Only a few species of yeast can reproduce by fission. In fission, cells grow to a certain size and divide into two equal cells. Sexual reproduction of yeasts involves the formation of a zygote (a diploid cell) from fusion of two haploid cells, each having a single set of chromosomes. The nucleus of the diploid cells divides several times to form ascospores. Each ascospore eventually becomes a new haploid cell and may reproduce by budding and fission. The life cycle of a typical yeast cell is presented in Figure 2.6.
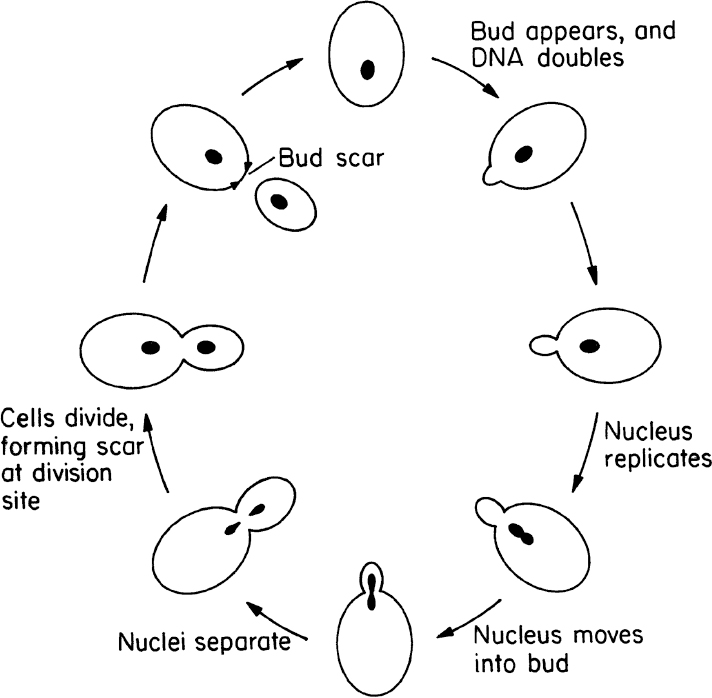
Figure 2.6. Cell-division cycle of a typical yeast, Saccharomyces cerevisiae. (With permission, from T. D. Brock, D. W. Smith, and M. T. Madigan, Biology of Microorganisms, 4th ed., Prentice Hall, Englewood Cliffs, NJ, 1984, p. 80.)
The classification of yeasts is based on reproductive modes (e.g., budding or fission) and the nutritional requirements of cells. The most widely used yeast, Saccharomyces cerevisiae, is used in alcohol formation under anaerobic conditions (e.g., in wine, beer, and whiskey making) and also for baker’s yeast production under aerobic conditions.
Molds are filamentous fungi and have a mycelial structure. The mycelium is a highly branched system of tubes that contains a mobile mass of cytoplasm with many nuclei. Long, thin filaments on the mycelium are called hyphae. Certain branches of mycelium may grow in the air, and asexual spores called conidia are formed on these aerial branches. Conidia are nearly spherical in structure and are often pigmented. Some molds reproduce by sexual means and form sexual spores. These spores provide resistance against heat, freezing, drying, and some chemical agents. Both sexual and asexual spores of molds can germinate and form new hyphae. Figure 2.7 describes the structure and asexual reproduction of molds.
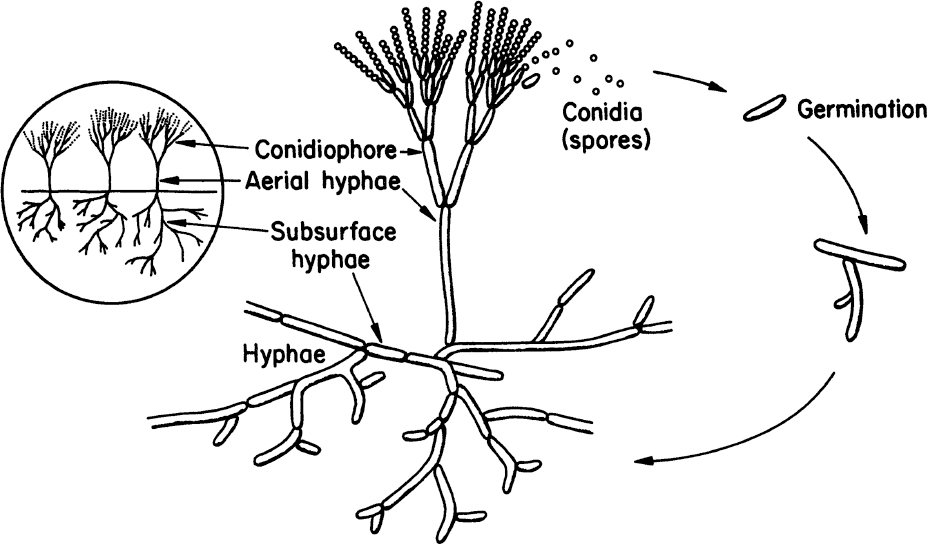
Figure 2.7. Structure and asexual reproduction of molds. (Thomas D. Brock, Katherine M. Brock, and David M. Ward, Basic Microbiology with Applications, 3d ed., © 1986. Reprinted and electronically reproduced by permission of Pearson Education, Inc., New York, NY.)
Molds usually form long, highly branched cells and easily grow on moist, solid nutrient surfaces. The typical size of a filamentous form of mold is 5 to 20 μm. When grown in submerged culture, molds often form cell aggregates and pellets. The typical size of a mold pellet varies between 50 μm and 1 mm, depending on the type of mold and growth conditions. Pellet formation can cause some nutrient-transfer (mainly oxygen) problems inside the pellet. However, pellet formation reduces broth viscosity, which can improve bulk oxygen transfer.
On the basis of their mode of sexual reproduction, fungi are grouped in four classes:
• The phycomycetes are algae-like fungi; however, they do not possess chlorophyll and cannot photosynthesize. Aquatic and terrestrial molds belong to this category.
• The ascomycetes form sexual spores called ascospores, which are contained within a sac (a capsule structure). Some molds of the genera Neurospora and Aspergillus and yeasts belong to this category.
• The basidiomycetes reproduce by basidiospores, which are extended from the stalks of specialized cells called the basidia. Mushrooms are basidiomycetes.
• The deuteromycetes (Fungi imperfecti) cannot reproduce by sexual means. Only asexually reproducing molds belong to this category. Some pathogenic fungi, such as Trichophyton, which causes athlete’s foot, belong to the deuteromycetes.
Molds are used for the production of citric acid (Aspergillus niger) and many antibiotics, such as penicillin (Penicillium chrysogenum). Mold fermentations make up a large fraction of the fermentation industry.
Algae are usually unicellular organisms. However, some plantlike multicellular structures are present in both marine and fresh waters. All algae are photosynthetic and contain chloroplasts, which normally impart a green color to the organisms. The chloroplasts are the sites of chlorophyll pigments and are responsible for photosynthesis. The size of a typical unicellular alga is 10 to 30 μm. Multicellular algae sometimes form a branched or unbranched filamentous structure. Some algae contain silica or calcium carbonate in their cell wall. Diatoms containing silica in their cell wall are used as filter aids in industry. Some algae, such as Chlorella, Scenedesmus, Spirulina, and Dunaliella, are used for wastewater treatment with simultaneous single-cell protein production. Certain gelling agents, such as agar and alginic acid, are obtained from marine algae and seaweeds. Some algae are brown or red because of the presence of other pigments. Certain microalgae species accumulate high levels of oil (>30%) and are of commercial interest because they may be used to provide a renewable platform for production of biodiesel.
Protozoa are unicellular, motile, relatively large (5 to 50 μm) eucaryotic cells that lack cell walls. Protozoa usually obtain food by ingesting other small organisms, such as bacteria, or other food particles. Protozoa are usually uninucleate and reproduce by sexual or asexual means. They are classified on the basis of their motion. The amoebae move by ameboid motion, whereby the cytoplasm of the cell flows forward to form a pseudopodium (false foot), and the rest of the cell flows toward this lobe. The flagellates move using their flagella. Trypanosomes move by flagella and cause a number of diseases in humans. The ciliates move by motion of a large number of small appendages on the cell surface called cilia. The sporozoans are nonmotile and contain members that are human and animal parasites. These protozoa do not engulf food particles but absorb dissolved food components through their membranes. Protozoa cause some diseases, such as malaria and dysentery. Protozoa may have a beneficial role in removing bacteria from wastewater in biological wastewater treatment processes and helping to obtain clean effluents. Microscopic pictures of some protozoa are presented in Figure 2.8.
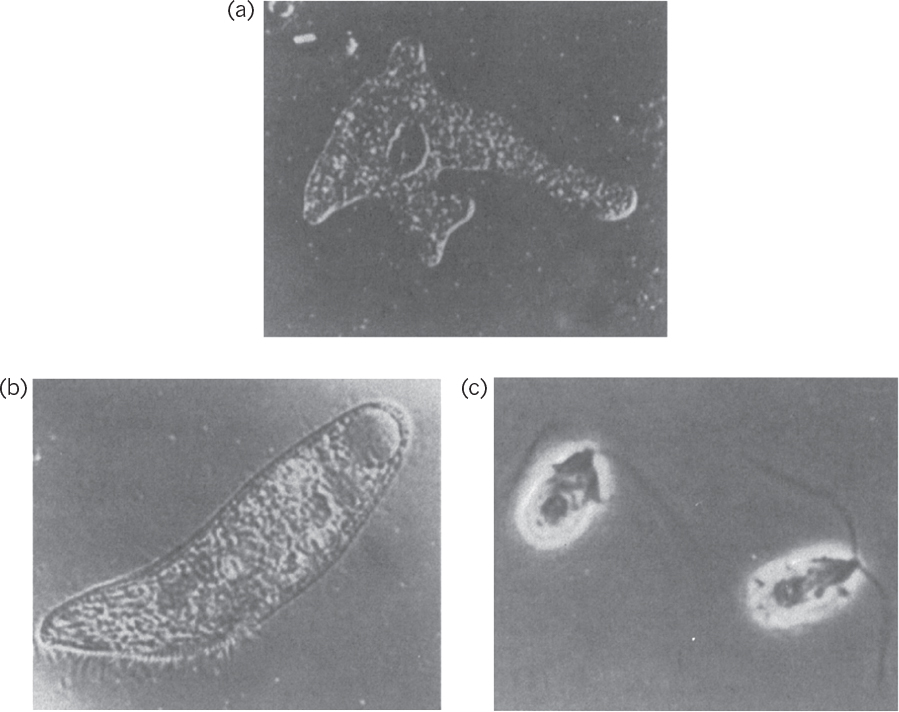
Figure 2.8. Microscopic images of protozoa. (a) An amoeba, Amoeba proteus. Magnification, 125X. (b) A ciliate, Blepharisma. Magnification, 120X. (c) A flagellate, Dunaliella. Magnification, 1900X. (Thomas D. Brock, Katherine M. Brock, and David M. Ward, Basic Microbiology with Applications, 3d ed., © 1986. Reprinted and electronically reproduced by permission of Pearson Education, Inc., New York, NY.)
2.2. Cell Construction
Living cells are composed of high-molecular-weight polymeric compounds such as proteins, nucleic acids, polysaccharides, lipids, and other storage materials (fats, polyhydroxybutyrate, glycogen). These biopolymers constitute the major structural elements of living cells. For example, a typical bacterial cell wall contains polysaccharides, proteins, and lipids; cell cytoplasm contains proteins mostly in the form of enzymes; in eucaryotes, the cell nucleus contains nucleic acids mostly in the form of DNA. In addition to these biopolymers, cells contain metabolites in the form of inorganic salts (e.g., ![]() ,
, ![]() , K+, Ca2+, Na+,
, K+, Ca2+, Na+, ![]() ), metabolic intermediates (e.g., pyruvate, acetate), and vitamins. The elemental composition of a typical bacterial cell is 50% carbon, 20% oxygen, 14% nitrogen, 8% hydrogen, 3% phosphorus, and 1% sulfur, with small amounts of K+, Na+, Ca2+, Mg2+, Cl–, and vitamins.
), metabolic intermediates (e.g., pyruvate, acetate), and vitamins. The elemental composition of a typical bacterial cell is 50% carbon, 20% oxygen, 14% nitrogen, 8% hydrogen, 3% phosphorus, and 1% sulfur, with small amounts of K+, Na+, Ca2+, Mg2+, Cl–, and vitamins.
The cellular macromolecules are functional only when in the proper three-dimensional configuration. The interaction among them is very complicated. Each macromolecule is part of an intracellular organelle and functions in its unique microenvironment. Information transfer from one organelle to another (e.g., from nucleus to ribosomes) is mediated by special molecules (e.g., messenger RNA). Most of the enzymes and metabolic intermediates are present in cytoplasm. However, other organelles, such as mitochondria, contain enzymes and other metabolites. A living cell can be visualized as a very complex reactor in which more than 2000 reactions take place. These reactions (metabolic pathways) are interrelated and are controlled in a complicated fashion.
Despite all their complexity, an understanding of biological systems can be simplified by analyzing the system at several different levels: molecular (molecular biology, biochemistry); cellular (cell biology, microbiology); population (microbiology, ecology); and production (bioprocess engineering). An emergent interdisciplinary field known as systems biology seeks to understand this complexity using a holistic approach (holism instead of the more traditional reductionism) to study the interactions within biological systems. This section is devoted mainly to the structure and function of biological molecules.
2.2.1. Amino Acids and Proteins
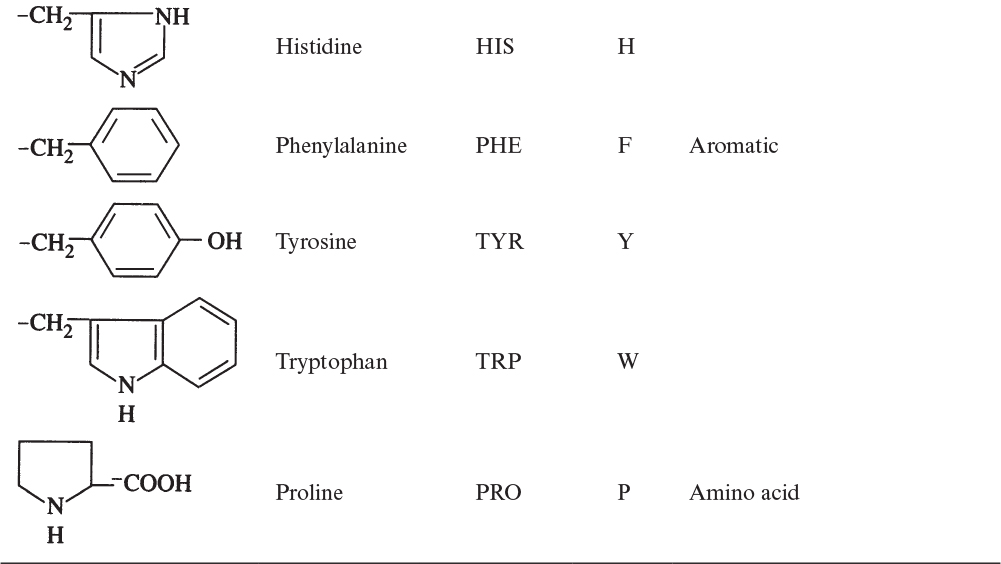
Proteins are the most abundant organic molecules in living cells, constituting 40% to 70% of their dry weight. Proteins are polymers built from amino acid monomers. Proteins typically have molecular weights (MWs) of 6000 to several hundred thousand. The α-amino acids are the building blocks of proteins and contain at least one carboxyl group and one α-amino group, but they differ from each other in the structure of their R groups or side chains:
Although the sequence of amino acids determines a protein’s primary structure, the secondary and tertiary structures are determined by the weak interactions among the various side groups. The ultimate three-dimensional structure is critical to the biological activity of the protein. Two major types of protein conformation are fibrous proteins and globular proteins. Figure 2.9 depicts examples of fibrous and globular proteins. Proteins have diverse biological functions, which can be classified in five major categories:
• Structural (e.g., collagen, elastin, keratin)
• Catalytic (e.g., amylase, catalase, lysozyme, pepsin)
• Transport (e.g., aquaporin, dopamine transporter, hemoglobin, serum albumin)
• Regulatory (e.g., human growth hormone, insulin sigma factor, transcription factor)
• Protective (e.g., antibodies, thrombin)

Figure 2.9. Fibrous and globular proteins. (From Lehninger Principles of Biochemistry, 6th ed., by David L. Nelson et al., Copyright 2013. All rights reserved. Reprinted by permission of W. H. Freeman.)
Enzymes represent the largest class of proteins. Over 2000 different kinds of enzymes are known. Enzymes are highly specific in their function and have extraordinary catalytic power. Enzyme molecules contain an active site to which specific substrates are bound during catalysis. Some enzymes regulate cellular reactions and are thus called regulatory enzymes. Although most enzymes are globular proteins, some reside in cellular membranes and are known as integral membrane proteins.
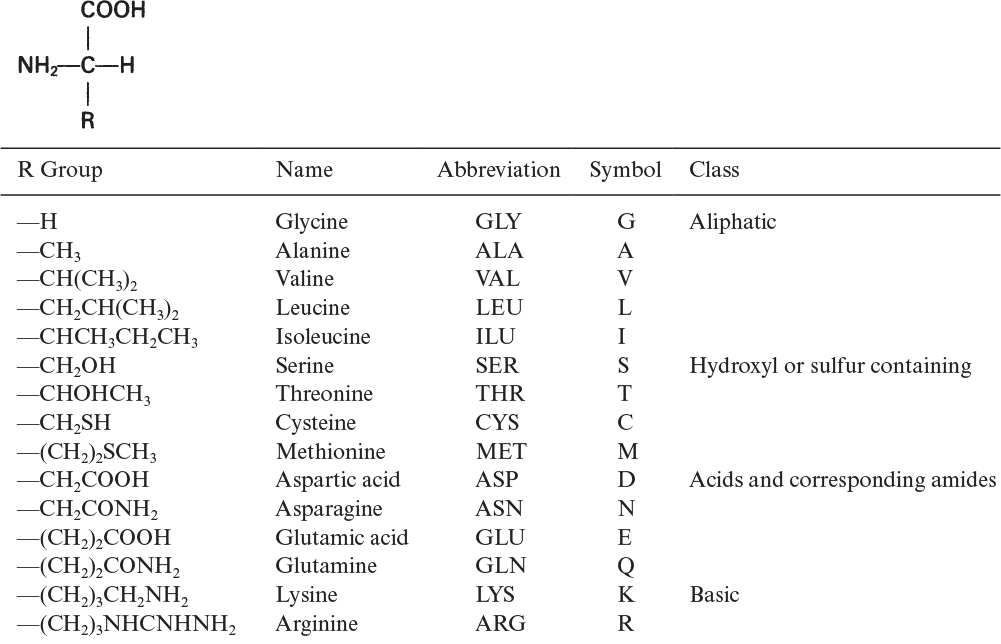
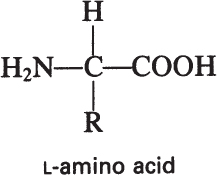
The building blocks of proteins are α-amino acids. There are 22 amino acids that can be naturally incorporated into polypeptides and are called proteinogenic or natural amino acids. Twenty of these are encoded by the universal genetic code, and the remaining two, selenocysteine and pyrrolysine (which are derivatives of cysteine and lysine), are incorporated into proteins by unique synthetic mechanisms. A list of the 20 common amino acids that are found in proteins is given in Table 2.4. Amino acids are named on the basis of the side (R) group attached to the α-carbon. Amino acids are optically active and occur in two isomeric forms:
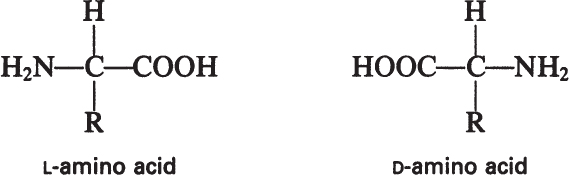
Only L-amino acids are found in proteins. D-amino acids are rare in nature; they are found in the cell walls of some microorganisms and in some antibiotics.
Amino acids have acidic (—COOH) and basic (—NH2) groups. The acidic group is neutral at low pH (—COOH) and negatively charged at high pH (—COO–). At intermediate pH values, an amino acid has positively and negatively charged groups, a dipolar molecule called a zwitterion:
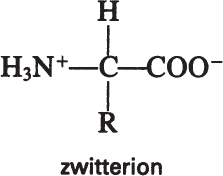
The pH value at which amino acids have no net charge is called the isoelectric point, which varies depending on the R group of amino acids. At its isoelectric point, an amino acid does not migrate under the influence of an electric field. Knowledge of the isoelectric point can be used in developing processes for protein purification.
Proteins are linear chains of covalently linked amino acids. The condensation reaction between two amino acids results in the formation of a peptide bond:
The peptide bond is planar. Peptides contain two or more amino acids linked by peptide bonds. Polypeptides usually contain fewer than 50 amino acids. Larger amino acid chains are called proteins. Many proteins contain organic and/or inorganic components other than amino acids. These components are called prosthetic groups, and the proteins containing prosthetic groups are named conjugated proteins. Hemoglobin is a conjugated protein and has four heme groups, which are iron-containing organometallic complexes.
The three-dimensional structure of proteins can be described at four levels:
• Primary structure: The primary structure of a protein is its linear sequence of amino acids. Each protein has not only a definite amino acid composition but also a unique sequence. The one-dimensional structure of proteins (the amino acid sequence) has a profound effect on the resulting three-dimensional structure and, therefore, on the function of proteins.
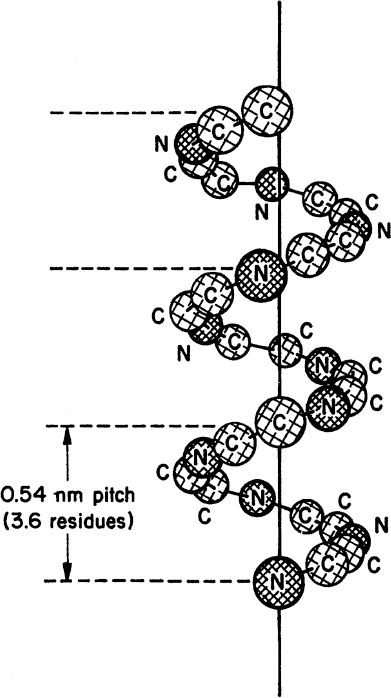
• Secondary structure: This structure is the means by which the polypeptide chain is extended and is a result of hydrogen bonding between residues not widely separated. Two major types of secondary structure are helixes and sheets. Helical structures can be either α-helical or triple helix. In an α-helical structure, hydrogen bonding can occur between the α-carboxyl group of one residue and the —NH group of its neighbor four units down the chain, as shown in Figure 2.10. The triple-helix structure present in collagen consists of three α-helixes intertwined in a superhelix. The triple-helix structure is rigid and stretch resistant. The α-helical structure can be easily disturbed, since H bonds are not highly stable. However, the sheet structure (β-pleated sheet) is more stable. The hydrogen bonds between parallel chains stabilize the sheet structure and provide resistance to stretching (Figure 2.11).
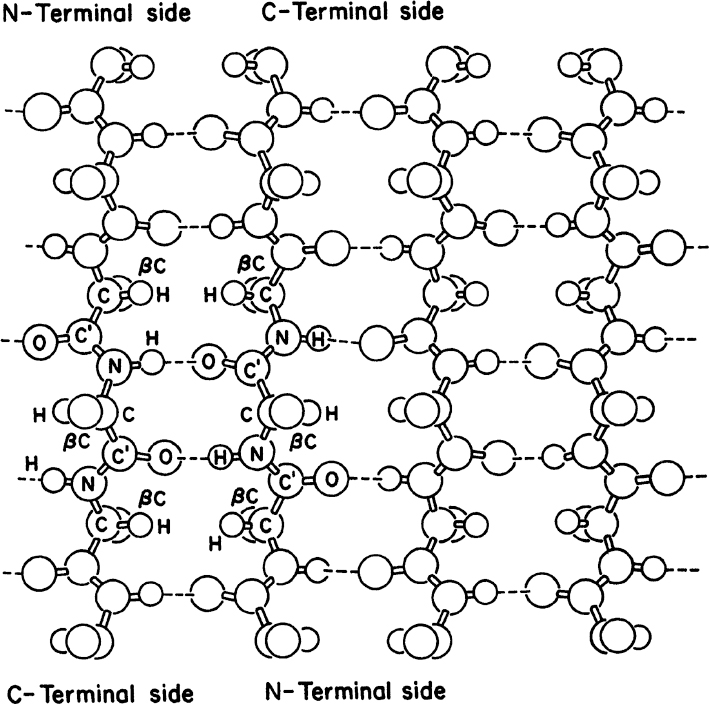
Figure 2.11. Representation of an antiparallel β-pleated sheet. Dashed lines indicate hydrogen bonds between strands.
• Tertiary structure: This structure is a result of interactions between R groups widely separated along the chain. The folding or bending of an amino acid chain, induced by interaction between R groups, determines the tertiary structure of proteins. R groups may interact by covalent, disulfide, or hydrogen bonds. Hydrophobic and hydrophilic interactions may also be present among R groups. A disulfide bond can cross-link two polypeptide chains (e.g., insulin). Disulfide bonds are also critical in proper chain folding, as shown in Figure 2.12. The tertiary structure of a protein has a profound effect on its function.

Figure 2.12. Structure of the enzyme ribonuclease. (a) Primary amino acid sequence, showing how sulfur–sulfur bonds between cysteine residues cause folding of the chain. (b) The three-dimensional structure of ribonuclease, showing how the macromolecule folds so that a site of enzymatic activity is formed, the active site. (Thomas D. Brock, Katherine M. Brock, and David M. Ward, Basic Microbiology with Applications, 3d ed., © 1986. Reprinted and electronically reproduced by permission of Pearson Education, Inc., New York, NY.)
• Quaternary structure: Only proteins with more than one polypeptide chain have quaternary structure. Interactions among polypeptide chains determine the quaternary structure (Figure 2.9). Hemoglobin has four subunits (oligomeric), and interaction among these subunits results in a quaternary structure. The forces between polypeptide chains can be disulfide bonds or other weak interactions. The subunit structure of enzymes has an important role in the control of their catalytic activity.
Antibodies or immunoglobulins are proteins that bind to particular molecules or portions of large molecules with a high degree of specificity. Antibody (Ab) molecules appear in the blood serum and in certain cells of a vertebrate in response to foreign macromolecules. The foreign macromolecule, called an antigen (Ag), is typically a pathogen, such as a bacterium or virus, or their constituents (protein, lipid, polysaccharide). A specific antibody molecule can combine with the antigen to form an antibody–antigen complex. Complex formation between antibodies and antigens is a key facet of the humoral immune response to a foreign invader. This interaction can be exploited for medical and industrial applications. For example, natural and engineered antibodies have emerged as a powerful modality for treating various human diseases in a strategy known as immunotherapy. One type of engineered antibody, known as an antibody-drug conjugate (ADC), has become a key element in the delivery of some anticancer drugs. In addition to their clinical significance, antibodies are important industrial products for use in diagnostic kits and protein separation schemes. Taken together, antibodies have emerged as one of the most important products of biotechnology.
Antibody and antigen molecules interact by spatial complementarity (lock and key). That is, antibodies have binding sites that are specific for and complementary to the structural features of the antigen’s epitopes, surface features arranged discontinuously along the antigen that are recognized by the antibody. Antibody molecules usually have two binding sites and can form a three-dimensional lattice of alternating antigen and antibody molecules. This complex precipitates from the serum and is called precipitin. Antibodies are highly specific for the foreign macromolecules that induce their formation.
The five major classes of immunoglobulins in human blood plasma are IgG, IgA, IgD, IgM, and IgE, of which the IgG globulins are the most abundant and the best understood. Immunoglobulin MWs are about 150 kilodaltons (kD); however, IgM forms a pentamer that has a MW of 900 kD, while the secreted form of IgA (sIgA) is composed of two to four IgA monomers linked by two additional chains and has a MW of 385 kD. A dalton is a unit of mass equivalent to a hydrogen atom. Immunoglobulins have four polypeptide chains: two heavy (H) chains (about 430 amino acids) and two light (L) chains (about 214 amino acids). These chains are linked together by disulfide bonds into a Y-shaped, flexible structure (Figure 2.13). The heavy chains contain a covalently bound oligosaccharide component known as a glycan. Each chain has a region of constant amino acid sequence and a variable-sequence region. The antibody molecule has two binding sites for the antigen; the variable portions of the L and H chains contribute to these binding sites. The variable sections have hypervariable regions or complementarity determining regions (CDRs) in which the frequency of amino acid replacement is high.

Figure 2.13. Immunoglobulin G structure. (Thomas D. Brock, Katherine M. Brock, and David M. Ward, Basic Microbiology with Applications, 14th ed., © 2015. Reprinted and electronically reproduced by permission of Pearson Education, Inc., New York, NY.)
How cells develop and produce antibodies is a heavily studied research topic worldwide. Recent developments have also led to insights on how to impart novel activities to antibodies, including cell penetration, multiantigen targeting (e.g., bispecific antibodies), and catalytic functions (e.g., abzymes). Coupling new developments in protein engineering to antibodies promises the development of extremely specific catalytic agents that can be delivered to specific cells or tissues by design.
2.2.2. Carbohydrates: Mono- and Polysaccharides
Carbohydrates play key roles as structural and storage compounds in cells. They also appear to play critical roles in modulating some aspects of chemical signaling in animals and plants. Carbohydrates are represented by the general formula (CH2O)n, where n ≥ 3, and are synthesized through photosynthesis.
The gases CO2 and H2O are converted through photosynthesis into sugars in the presence of sunlight and are then polymerized to yield polysaccharides, such as cellulose or starch.
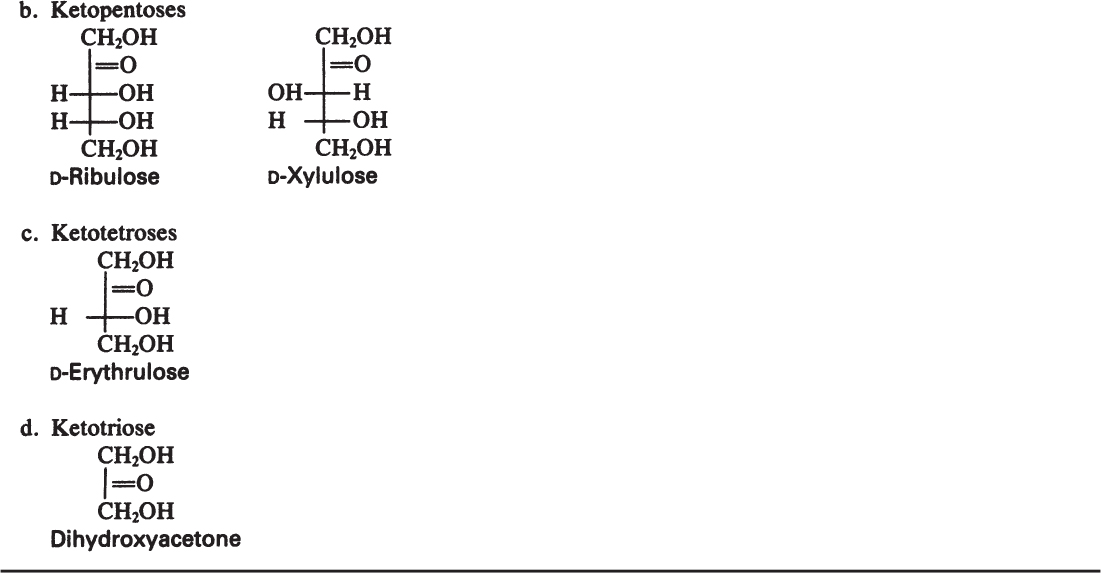
Monosaccharides are the smallest carbohydrates and contain three to nine carbon atoms. Common monosaccharides are presented in Table 2.5. Common monosaccharides are either aldehydes or ketones. For example, glucose is an aldohexose. Glucose may be present in the form of a linear or ring structure. In solution, D-glucose is in the form of a ring (pyranose) structure. The L-form plays a minor role in biological systems.

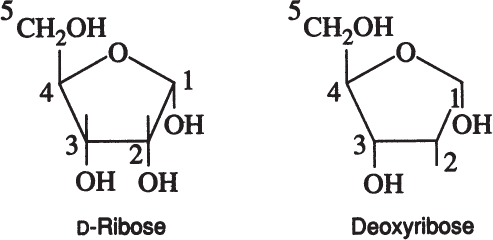
A particularly important group of monosaccharides are D-ribose and deoxyribose. These are five carbon ring-structured sugar molecules and are essential components of DNA and RNA.
Disaccharides are formed by the condensation of two monosaccharides. For example, maltose is formed by the condensation of two glucose molecules via an α-1,4 glycosidic linkage.

Sucrose is a disaccharide of α-D-glucose and β-D-fructose. Lactose is a disaccharide of β-D-glucose and β-D-galactose.
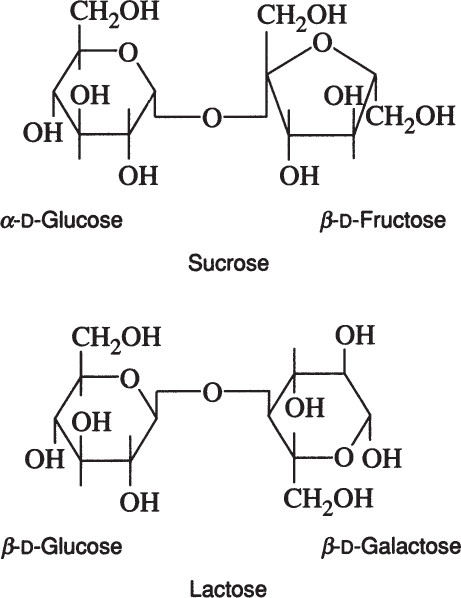
Lactose is found in milk and whey, and sucrose is the major sugar in photosynthetic plants. Whey utilization remains an important biotechnological challenge, and sucrose is often a major component in artificial growth media.
Polysaccharides are formed by the condensation of more than two monosaccharides by glycosidic bonds and can be assembled in linear and branched configurations. The polysaccharide processing industry makes extensive use of enzymatic processing and biochemical engineering.
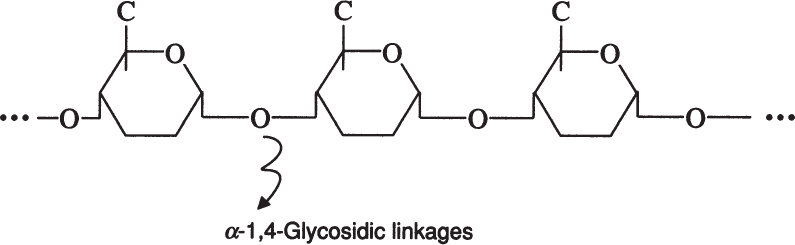
Amylose is a straight chain of glucose molecules linked by α-1,4 glycosidic linkages. The MW of amylose is between several thousand and one-half million daltons. Amylose is water insoluble and constitutes about 20% of starch.
Amylopectin is a branched chain of D-glucose molecules. Branching occurs between the glycosidic —OH of one chain and the —6 carbon of another glucose, which is called an α-1,6 glycosidic linkage.
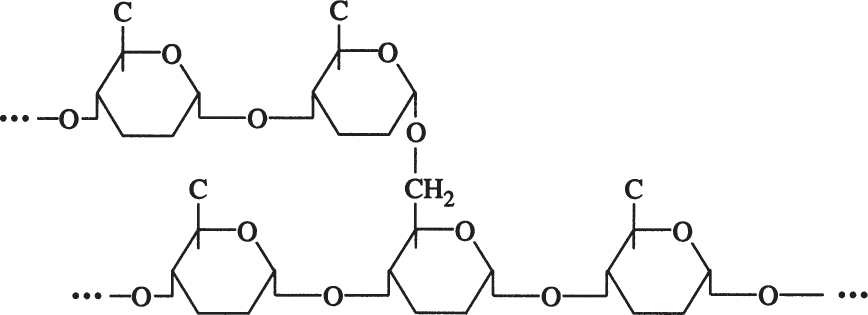
Amylopectin molecules are much larger than those of amylose, with a MW of 1 to 2 million daltons. Amylopectin is water soluble. Partial hydrolysis of starch (acidic or enzymatic) yields glucose, maltose, and dextrins, which are branched sections of amylopectin. Dextrins are used as thickeners.
Glycogen is a branched chain of glucose molecules that resembles amylopectin. Glycogen is highly branched and contains about 12 glucose units in straight-chain segments. The MW of a typical glycogen molecule is less than 5 × 106 daltons.
Cellulose is a long, unbranched chain of D-glucose with a MW between 50,000 and 1 million daltons. The linkage between glucose monomers in cellulose is a β-1,4 glycosidic linkage.
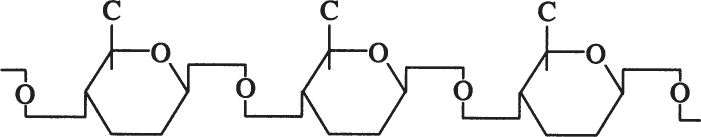
The β-1,4 glycosidic bond is resistant to enzymatic hydrolysis. Only a few microorganisms can hydrolyze β-1,4 glycosidic bonds of cellulose. α-1,4 glycosidic bonds in starch or glycogen are relatively easy to break by enzymatic or acid hydrolysis. Efficient cellulose hydrolysis remains one of the most challenging problems in attempts to convert cellulosic materials into valuable fuels or chemicals.
Polysaccharides can exist in a free form or as attachments to other biomolecules. As conjugates, polysaccharides or glycans add an additional layer of information to biomolecules and have diverse biological roles such as regulating protein structure and function and altering cell surface properties. The process whereby a glycan is enzymatically attached to a hydroxyl or other functional group of another biomolecule (e.g., protein, lipid) is referred to as glycosylation. When attached to proteins, glycans can influence folding, stability, molecular interactions, and quality control. Over half of all eucaryotic proteins are predicted to be glycosylated, although yeasts have fewer total glycoproteins than multicellular eucaryotes. Not surprisingly, approximately 70% of therapeutic proteins either approved or in clinical and preclinical development are glycoproteins.
2.2.3. Lipids, Fats, and Steroids
Lipids are hydrophobic biological compounds that are insoluble in water but soluble in nonpolar solvents such as benzene, chloroform, and ether. They are usually present in the nonaqueous biological phases, such as plasma membranes. Fats are lipids that can serve as biological fuel-storage molecules. Lipoproteins and lipopolysaccharides are other types of lipids, which appear in the biological membranes of cells. Cells can alter the mix of lipids in their membranes to compensate (at least partially) for changes in temperature or to increase their tolerance to the presence of chemical agents such as ethanol.
The major component in most lipids is fatty acids, which consist of a straight chain of hydrocarbon (hydrophobic) groups, with a carboxyl group (hydrophilic) at the end. A typical fatty acid can be represented as follows:
CH3 —(CH2)n —COOH
The value of n is typically between 12 and 20. Unsaturated fatty acids contain double —C = C— bonds, such as oleic acid:
A list of common fatty acids is presented in Table 2.6. The hydrocarbon chain of a fatty acid is hydrophobic (water insoluble), but the carboxyl group is hydrophilic (water soluble).
Fats are esters of fatty acids with glycerol. The formation of a fat molecule can be represented by the following reaction:
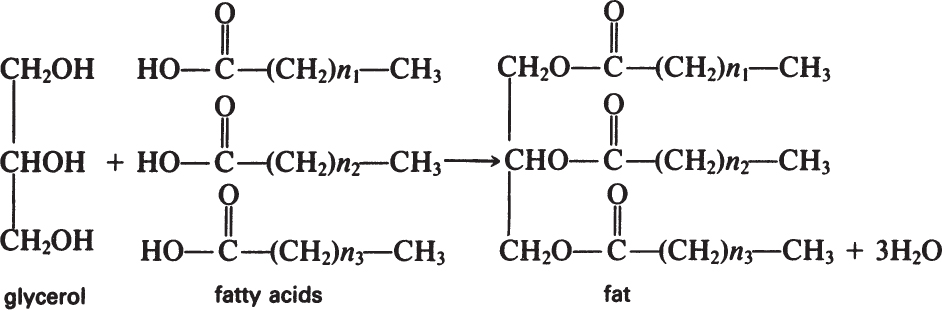
Phosphoglycerides have similar structures to fats, the only difference being that phosphoric acid replaces a fatty acid and is esterified at one end to glycerol.
Membranes with selective permeability are key to life. Cells must control the entry and exit of molecules. Phospholipids are key components, but membranes contain large amounts of proteins. Biological membranes are based on a lipid bilayer. The hydrophobic tails of the phospholipids associate with each other in the core of the membrane. The hydrophilic heads form the outsides of the membrane and associate with the aqueous cytosol or the aqueous extracellular fluid. Some proteins span across the membrane (integral membrane proteins), whereas others are attached to one of the surfaces (peripheral membrane proteins). Membranes are dynamic structures, and lipids and proteins can diffuse rapidly. Typical membrane phospholipids include phosphatidylcholine, phosphatidylserine, phosphatidyl glycerol, and phosphatidyl inositol.
Another class of lipids of increasing technological importance is the polyhydroxyalkanoates (PHA). In particular, polyhydroxybutyrate (PHB) is a good example. It can be used to form a clear, biodegradable polymeric sheet. Polymers with a variety of PHAs are being commercially developed. In some cells, PHB is formed as a storage product.
Steroids can also be classified as lipids. Naturally occurring steroids are hormones that are important regulators of animal development and metabolism at very low concentrations (e.g., 10–8 M). A well-known steroid, cholesterol, is present in membranes of animal tissues. Figure 2.14 depicts the structures of some important steroids. Cortisone is an anti-inflammatory used to treat rheumatoid arthritis and some skin diseases. Derivatives of estrogens and progesterone are used as contraceptives. The commercial production of steroids is very important and depends on microbial conversions. Because of the large number of asymmetric centers, the total synthesis of steroids is difficult. Plants provide a source of abundant lipid precursors for these steroids, but the highly specific hydroxylation of these substrates at positions 11 (and 16) or dehydrogenations at position 1 are necessary to convert the precursors into compounds similar to those made in the adrenal gland. Such conversions cannot be done easily with chemical means and is done commercially using microbes that contain enzymes mediating specific hydroxylations or dehydrogenations.

Figure 2.14. Examples of important steroids. The basic numbering of the carbon atoms in these molecules is also shown.
2.2.4. Nucleic Acids, RNA, and DNA
Nucleic acids play a central role in reproduction of living cells. Deoxyribonucleic acid (DNA) stores and preserves genetic information. Ribonucleic acid (RNA) plays a central role in protein synthesis. Both DNA and RNA are large polymers made of their corresponding nucleotides.
Nucleotides are the building blocks of DNA and RNA and also serve as molecules to store energy and reducing power. The three major components in all nucleotides are phosphoric acid, pentose (ribose or deoxyribose), and a base (purine or pyrimidine). Figure 2.15 depicts the structure of nucleotides and purine-pyrimidine bases. Two major purines present in nucleotides are adenine (A) and guanine (G), and three major pyrimidines are thymine (T), cytosine (C), and uracil (U). DNA contains A, T, G, and C, and RNA contains A, U, G, and C as bases. It is the base sequence in DNA that carries genetic information for protein synthesis. In Chapter 4, “How Cells Work,” and Chapter 8, “How Cellular Information Is Altered,” we discuss how this information is expressed and passed on from one generation to another.

Figure 2.15. (a) General structure of ribonucleotides and deoxyribonucleotides. (b) Five nitrogenous bases found in DNA and RNA. (With permission, from J. E. Bailey and D. F. Ollis, Biochemical Engineering Fundamentals, 2d ed., McGraw-Hill, New York, 1986, p. 43.)
In Chapter 5, “Major Metabolic Pathways,” we discuss further the role of nucleotides in cellular energetics. The triphosphates of adenosine and to a lesser extent guanosine are the primary energy currency of the cell. The phosphate bonds in ATP (adenosine triphosphate) and GTP (guanosine triphosphate) are high-energy bonds. The formation of these phosphate bonds or their hydrolysis is the primary means by which cellular energy is stored or used. For example, the synthesis of a compound that is thermodynamically unfavorable can be coupled to hydrolysis of ATP to ADP (adenosine diphosphate) or AMP (adenosine monophosphate). The coupled reaction can proceed to a much greater extent, since the free-energy change becomes much more negative. In reactions that release energy (e.g., oxidation of a sugar), the energy is “captured” and stored by the formation of a phosphate bond in a coupled reaction where ADP is converted into ATP.
In addition to using ATP to store energy, the cell stores and releases hydrogen atoms from biological oxidation-reduction reactions by using nucleotide derivatives. The two most common carriers of reducing power are nicotinamide adenine dinucleotide (NAD) and nicotinamide adenine dinucleotide phosphate (NADP).
In addition to this important role in cellular energetics, the nucleotides are important monomers. The polynucleotides (DNA and RNA) are formed by the condensation of nucleotides. The nucleotides are linked together between the 3′ and 5′ carbons’ successive sugar rings by phosphodiester bonds. The structures of DNA and RNA are illustrated in Figure 2.16.

Figure 2.16. Structure of DNA and RNA molecules. Phosphodiester bonds are formed between 3′ and 5′ carbon atoms. (From Lehninger Principles of Biochemistry, 6th ed., by David L. Nelson et al., Copyright 2013. All rights reserved. Reprinted by permission of W. H. Freeman.)
DNA is a very large, threadlike macromolecule (MW, 2 × 109 D in E. coli) and has a double-helical three-dimensional structure. The sequence of bases (purines and pyrimidines) in DNA carries genetic information, whereas sugar and phosphate groups perform a structural role. The base sequence of DNA is written in the 5′ → 3′ direction, such as pAGCT. The double-helical structure of DNA is depicted in Figure 2.17. In this structure, two helical polynucleotide chains are coiled around a common axis to form a double-helical DNA, and the chains run in opposite directions, 5′ → 3′ and 3′ → 5′. The main features of double-helical DNA structures are as follows:
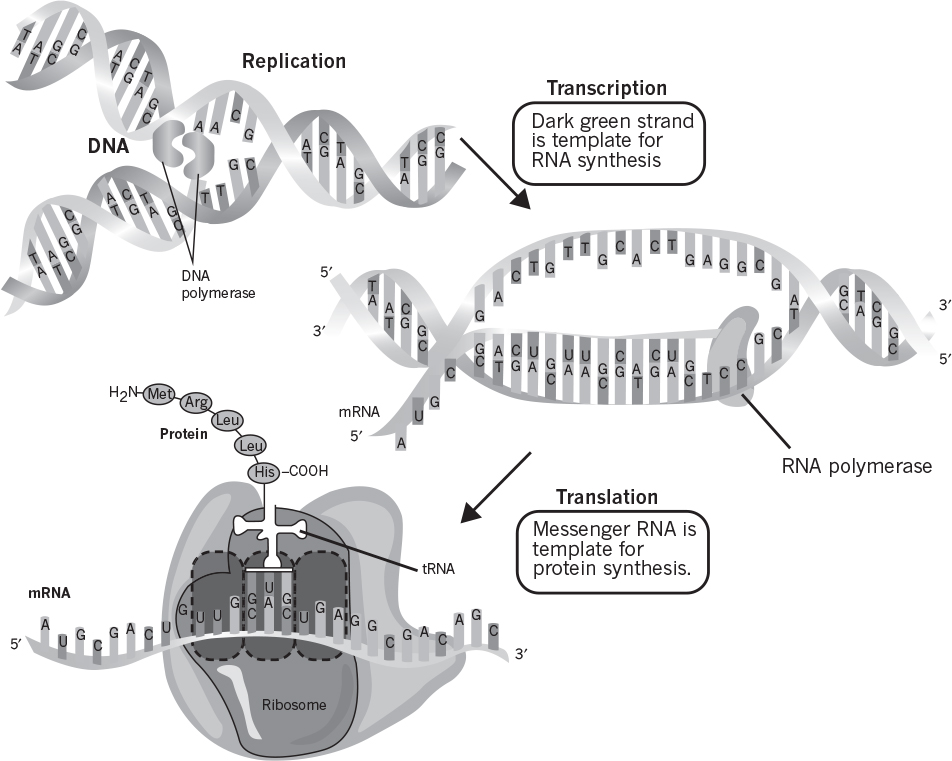
Figure 2.17. Double-helical structure of DNA, showing overall process of replication by complementary base pairing. (Thomas D. Brock, Katherine M. Brock, and David M. Ward, Basic Microbiology with Applications, 14th ed., © 2015. Reprinted and electronically reproduced by permission of Pearson Education, Inc., New York, NY.)
• The phosphate and deoxyribose units are on the outer surface, but the bases point toward the chain center. The planes of the bases are perpendicular to the helix axis.
• The diameter of the helix is 2 nm. The helical structure repeats after 10 residues on each chain, at an interval of 3.4 nm.
• The two chains are held together by hydrogen bonding between pairs of bases. Adenine is always paired with thymine (two H bonds); guanine is always paired with cytosine (three H bonds). This feature is essential to the genetic role of DNA.
• The sequence of bases along a polynucleotide is not restricted in any way, although each strand must be complementary to the other. The precise sequence of bases carries the genetic information.
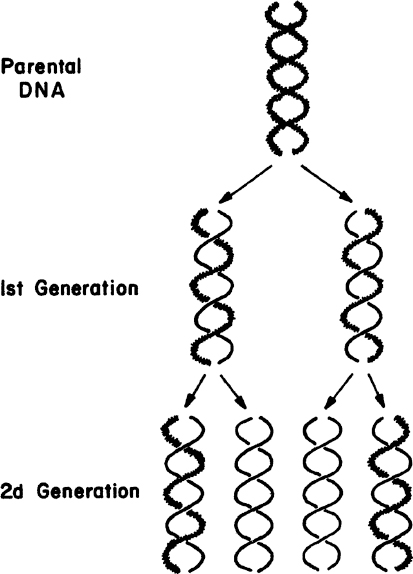
The large number of H bonds formed between base pairs provides molecular stabilization. Regeneration of DNA from original DNA segments is known as DNA replication. When DNA segments are replicated, one strand of the new DNA segment comes directly from the parent DNA, and the other strand is newly synthesized using the parent DNA segment as a template. Therefore, DNA replication is semiconservative, as depicted in Figure 2.18. The replication of DNA is discussed in more detail in Chapter 4.
Some cells contain circular DNA segments in the cytoplasm that are called plasmids. (Linear rather than circular plasmids can be found in some yeasts and other organisms.) Plasmids are nonchromosomal, autonomous, self-replicating DNA segments. Plasmids are easily moved in and out of the cells and are often used for genetic engineering, which is discussed in more detail in Chapter 8. Naturally occurring plasmids can encode factors that protect cells from antibiotics or harmful chemicals.
The major function of DNA is to carry genetic information in its base sequence. The genetic information in DNA is transcribed by RNA molecules and translated in protein synthesis. The templates for RNA synthesis are DNA molecules, and RNA molecules are the templates for protein synthesis. The formation of RNA molecules from DNA is known as DNA transcription, and the formation of peptides and proteins from RNA is called translation.
Certain RNA molecules function as the genetic information-carrying intermediates in protein synthesis (messenger, mRNA), whereas other RNA molecules (transfer [tRNA] and ribosomal [rRNA]) are part of the machinery of protein synthesis. The rRNA is located in ribosomes, which are small particles made of protein and RNA. Ribosomes are cytoplasmic organelles (usually attached on the inner surfaces of endoplasmic reticulum in eucaryotes) and are the sites of protein synthesis.
RNA is a long, unbranched macromolecule consisting of nucleotides joined by 3′ → 5′ phosphodiester bonds. An RNA molecule may contain from 70 to several thousand nucleotides. RNA molecules are usually single stranded, except some viral RNA. However, certain RNA molecules contain regions of double-helical structure, like hairpin loops. Figure 2.19 describes the cloverleaf structure of tRNA. In double-helical regions of tRNA, A pairs with U and G pairs with C. The RNA content of cells is usually two to six times higher than the DNA content.
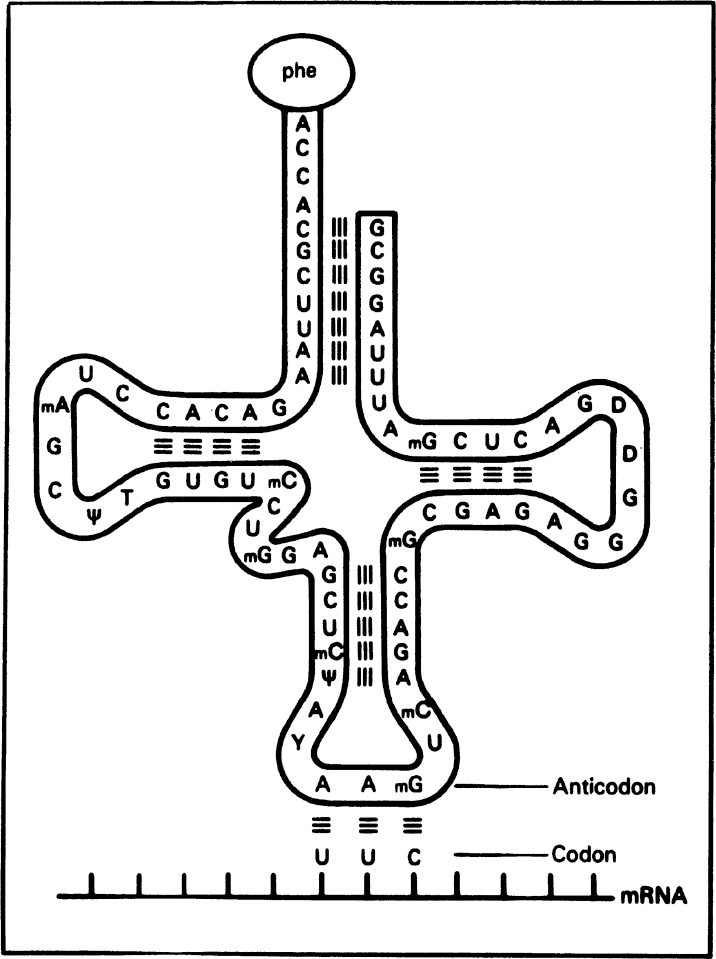
Figure 2.19. The structure of a transfer RNA (tRNA) molecule and the manner in which the anticodon of tRNA associates with the codon on messenger RNA (mRNA) by complementary base pairing. The amino acid corresponding to this codon (UUC) is phenylalanine, which is bound to the opposite end of the tRNA molecule. Many tRNA molecules contain unusual bases, such as methyl cytosine (mC) and pseudouridine (ψ). (Thomas D. Brock, Katherine M. Brock, and David M. Ward, Basic Microbiology with Applications, 3d ed., © 1986. Reprinted and electronically reproduced by permission of Pearson Education, Inc., New York, NY.)
It is important to note that an organism’s DNA includes sequences that code for proteins (coding sequences) as well as sequences that do not (noncoding DNA; originally termed “junk” DNA). Some of the noncoding DNA is transcribed into the noncoding RNA (ncRNA) molecules discussed earlier (e.g., tRNAs, rRNAs) as well as into regulatory RNAs. Two types of important small regulatory RNAs are microRNAs (miRNAs) and small interfering RNAs (siRNAs), which can bind to other specific mRNA molecules and either increase or decrease their activity, for example, by preventing an mRNA from producing a protein. Likewise, long ncRNAs (lncRNAs) play a functional role in regulating gene transcription as well as posttranscriptional mRNA processing.
Other functions of noncoding DNA include mediating structural organization of the chromatin within the nucleus and regulating the transcription and translation of protein-coding sequences by defining the location where transcription factors can attach and control transcription of the genetic code from DNA to mRNA.
Let us summarize the roles of each class of RNA species:
• Messenger RNA (mRNA) is synthesized on the chromosome and carries genetic information from the chromosome for synthesis of a particular protein to the ribosomes. The mRNA molecule is a large one with a short half-life.
• Transfer RNA (tRNA) is a relatively small and stable molecule that carries a specific amino acid from the cytoplasm to the site of protein synthesis on ribosomes. tRNAs contain 70 to 90 nucleotides and have a MW range of 23 to 28 kD. Each one of 20 amino acids has at least one corresponding tRNA.
• Ribosomal RNA (rRNA) is the major component of ribosomes, constituting nearly 65%. The remainder is various ribosomal proteins. Three distinct types of rRNAs present in the E. coli ribosome are specified as 23S, 16S, and 5S, respectively, on the basis of their sedimentation coefficients (determined in a centrifuge). The symbol S denotes a Svedberg unit. The MWs are 35 kD for 5S, 550 kD for 16S, and 1100 kD for 23S. These three rRNAs differ in their base sequences and ratios. Eucaryotic cells have larger ribosomes and four different types of rRNAs: 5S, 7S, 18S, and 28S. rRNAs make up a large fraction of total RNA. In E. coli, about 85% of the total RNA is rRNA, while tRNA is about 12% and mRNA is 2% to 3%.
• Noncoding RNA (ncRNA) is any RNA molecule that is not translated into a protein. This class includes the highly abundant and functionally important tRNA and rRNA molecules. It also includes small regulatory RNAs such as miRNAs, siRNAs, and small nucleolar RNAs (snoRNAs) among others, as well as long ncRNAs (lncRNAs). The number of nucleotides for these RNAs is approximately 22 for miRNAs, 20 to 25 for siRNAs, 20 to 200 for snoRNAs, and longer than 200 for lncRNAs.
2.3. Cell Nutrients
A cell’s composition differs greatly from its environment. A cell must selectively remove desirable compounds from its extracellular environment and retain other compounds intracellularly. A semipermeable membrane is the key to this selectivity. Since the cell differs so greatly in composition from its environment, it must expend energy to maintain itself away from thermodynamic equilibrium. Thermodynamic equilibrium and death are equivalent for a cell.
All organisms except viruses contain large amounts of water (about 80%). About 50% of cellular dry weight is protein, and the proteins are largely enzymes (proteins that act as catalysts). The nucleic acid content (which contains the genetic code and machinery to make proteins) of cells varies from 10% to 20% of dry weight. However, viruses may contain nucleic acids up to 50% of their dry weight. Typically, the lipid content of most cells varies between 5% and 15% of dry weight. However, some cells accumulate PHB up to 90% of the total mass under certain culture conditions. In general, the intracellular composition of cells varies depending on the type and age of the cells and the composition of the nutrient media. Typical compositions for major groups of organisms are summarized in Table 2.7.
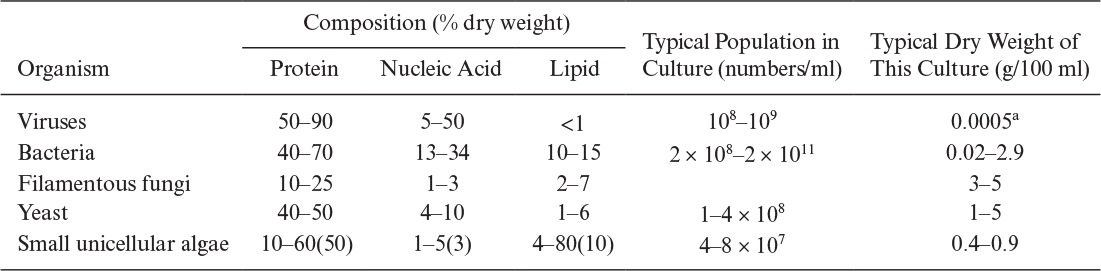
TABLE 2.7. Chemical Analyses, Dry Weights, and the Populations of Different Microorganisms Obtained in Culture
Most of the products formed by organisms are produced as a result of their response to environmental conditions, such as nutrients, growth hormones, and ions. The qualitative and quantitative nutritional requirements of cells need to be determined to optimize growth and product formation. Nutrients required by cells can be classified in two categories:
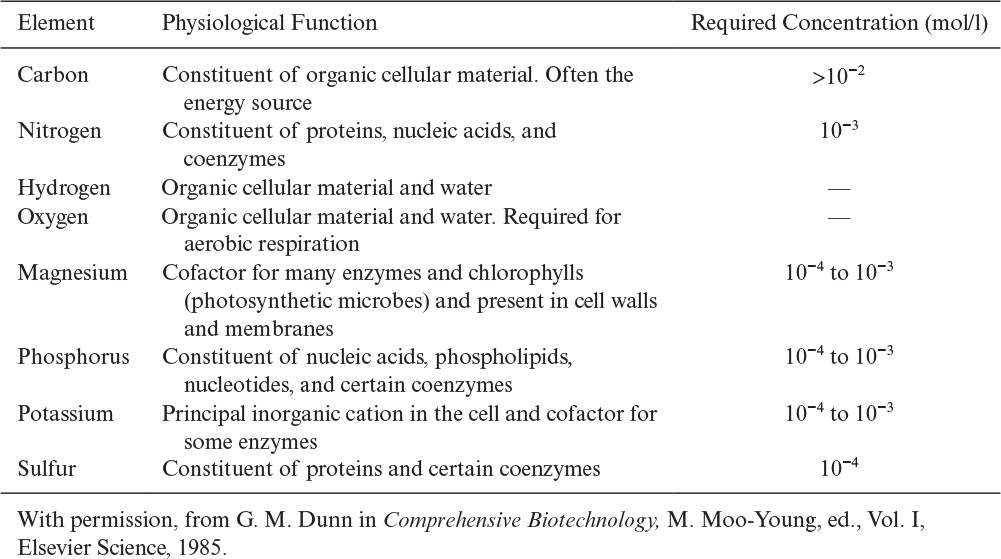
• Macronutrients are needed in concentrations larger than 10–4 M. Carbon, nitrogen, oxygen, hydrogen, sulfur, phosphorus, Mg2+, and K+ are major macronutrients.
• Micronutrients are needed in concentrations of less than 10–4 M. Trace elements such as Mo2+, Zn2+, Cu2+, Mn2+, Ca2+, Na+, vitamins, growth hormones, and metabolic precursors are micronutrients.
2.3.1. Macronutrients
Carbon compounds are major sources of cellular carbon and energy. Microorganisms are classified in two categories on the basis of their carbon source: heterotrophs and autotrophs. Heterotrophs use organic compounds such as carbohydrates, lipids, and hydrocarbons as a carbon and energy source. Autotrophs use carbon dioxide as a carbon source. Mixotrophs concomitantly grow under both autotrophic and heterotrophic conditions; however, autotrophic growth is stimulated by certain organic compounds. Facultative autotrophs normally grow under autotrophic conditions; however, they can grow under heterotrophic conditions in the absence of CO2 and inorganic energy sources. Chemoautotrophs utilize CO2 as a carbon source and obtain energy from the oxidation of inorganic compounds. Photoautotrophs use CO2 as a carbon source and utilize light as an energy source.
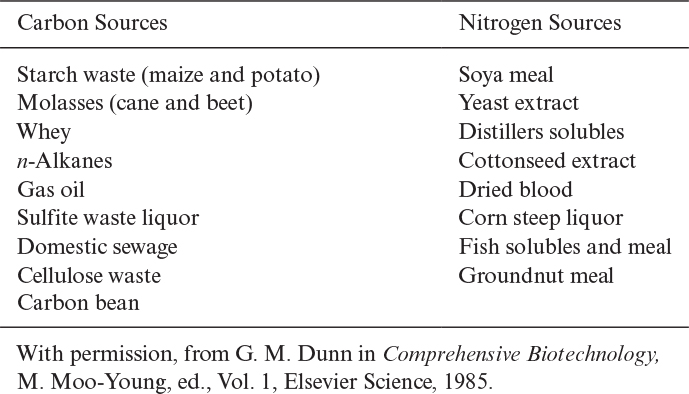
The most common carbon sources in industrial fermentations are molasses (sucrose), starch (glucose, dextrin), corn syrup, and waste sulfite liquor (glucose). In laboratory fermentations, glucose, sucrose, and fructose are the most common carbon sources. Methanol, ethanol, and methane also constitute cheap carbon sources for some fermentations. In aerobic fermentations, about 50% of substrate carbon is incorporated into cells and about 50% of it is used as an energy source. In anaerobic fermentations, a large fraction of substrate carbon is converted to products and a smaller fraction is converted to cell mass (less than 20%).
Nitrogen constitutes about 10% to 14% of cell dry weight. The most widely used nitrogen sources are ammonia or the ammonium salts (NH4Cl, [NH4]2SO4, NH4NO3), proteins, peptides, and amino acids. Nitrogen is incorporated into cell mass in the form of proteins and nucleic acids. Some organisms such as Azotobacter species and many species of cyanobacteria fix nitrogen from the atmosphere to form ammonium. Urea may also be used as a nitrogen source by some organisms. Organic nitrogen sources such as yeast extract and peptone are expensive compared to ammonium salts. Some carbon and nitrogen sources utilized by the fermentation industry are summarized in Table 2.8.
Oxygen is present in all organic cell components and cellular water and constitutes about 15% to 20% of the dry weight of cells. Molecular oxygen is required as a terminal electron acceptor in the aerobic metabolism of carbon compounds. Gaseous oxygen is introduced into growth media by sparging air or by surface aeration.
Hydrogen constitutes about 8% of cell dry weight and is derived primarily from carbon compounds, such as carbohydrates. Some bacteria, such as methanogens, can utilize hydrogen as an energy source.
Magnesium is a cofactor for some enzymes and is present in cell walls and membranes. Ribosomes specifically require Mg2+ ions. ATP molecules in the cytoplasm are usually bound to Mg2+. Magnesium is usually supplied as MgSO4 · 7H2O or MgCl2.
Phosphorus constitutes about 2% to 3% of cell dry weight and is present in nucleic acids and in the cell wall of some gram-positive bacteria such as teichoic acids. Some bacteria deposit polyphosphate granules. Inorganic phosphate salts, such as KH2PO4 and K2HPO4, are the most common phosphate salts. Glycerophosphates can also be used as organic phosphate sources. Phosphorus is a key element in the regulation of cell metabolism. The phosphate level in the media should be less than 1 mM for the formation of many secondary metabolites such as antibiotics.
Potassium is a cofactor for some enzymes and is required in carbohydrate metabolism. Cells tend to actively take up K+ and Mg2+ and exclude Na+ and Ca2+. The most commonly used potassium salts are K2HPO4, KH2PO4, and K3PO4.
Sulfur constitutes nearly 0.5% to 1% of cell dry weight and is present in proteins and some coenzymes. Sulfate salts such as (NH4)2SO4 are the most common sulfur source. Sulfur-containing amino acids can also be used as a sulfur source. Certain autotrophs utilize S2+ and S0 as energy sources.
Table 2.9 lists the eight major macronutrients and their physiological role.
2.3.2. Micronutrients
Trace elements are essential to microbial nutrition. Lack of essential trace elements increases the lag phase (the time from inoculation to active cell replication in batch culture) and may decrease the specific growth rate and the yield. Following are the three major categories of micronutrients:
• Commonly added: The most widely needed trace elements are Fe, Zn, and Mn. Iron (Fe) is present in ferredoxin and cytochrome and is an important cofactor. Iron also plays a regulatory role in some fermentation processes (e.g., iron deficiency is required for the excretion of riboflavin by Ashbya gossypii, and iron concentration regulates penicillin production by P. chrysogenum). Zinc (Zn) is a cofactor for some enzymes and also regulates some fermentations such as penicillin fermentation. Manganese (Mn) is also an enzyme cofactor and plays a role in the regulation of secondary metabolism and excretion of primary metabolites.
• Sometimes added: Trace elements needed under specific growth conditions are Cu, Co, Mo, Ca, Na, Cl, Ni, and Se. Copper (Cu) is present in certain respiratory-chain components and enzymes. Copper deficiency stimulates penicillin and citric acid production. Cobalt (Co) is present in corrinoid compounds such as vitamin B12. Propionic bacteria and certain methanogens require cobalt. Molybdenum (Mo) is a cofactor of nitrate reductase and nitrogenase and is required for growth on NO3 and N2 as the sole source of nitrogen. Calcium (Ca) is a cofactor for amylases and some proteases and is also present in some bacterial spores and in the cell walls of some cells, such as plant cells. Sodium (Na) is needed in trace amounts by some bacteria, especially by methanogens for ion balance. Sodium is important in the transport of charged species in eucaryotic cells. Chloride (Cl–) is needed by some halobacteria and marine microbes, which require Na+, too. Nickel (Ni) is required by some methanogens as a cofactor, and Selenium (Se) is required in formate metabolism of some organisms.
• Rarely added: Trace elements that are rarely required are B, Al, Si, Cr, V, Sn, Be, F, Ti, Ga, Ge, Br, Zr, W, Li, and I. These elements are required in concentrations of less than 10–6 M and are toxic at high concentrations, such as 10–4 M.
Some ions such as Mg2+, Fe3+, and ![]() may precipitate in nutrient medium and become unavailable to the cells. Chelating agents are used to form soluble compounds with the precipitating ions. Chelating agents have certain groups termed ligands that bind to metal ions to form soluble complexes. Major ligands are carboxyl (—COOH), amine (—NH2), and mercapto (—SH) groups. Citric acid, EDTA (ethylenediaminetetraacetic acid), polyphosphates, histidine, tyrosine, and cysteine are the most commonly used chelating agents. Na2-EDTA is the most common chelating agent. EDTA may remove some metal ion components of the cell wall, such as Ca2+, Mg2+, and Zn2+, and may cause cell-wall disintegration. Citric acid is metabolizable by some bacteria. Chelating agents are included in media in low concentrations (e.g., 1 mM).
may precipitate in nutrient medium and become unavailable to the cells. Chelating agents are used to form soluble compounds with the precipitating ions. Chelating agents have certain groups termed ligands that bind to metal ions to form soluble complexes. Major ligands are carboxyl (—COOH), amine (—NH2), and mercapto (—SH) groups. Citric acid, EDTA (ethylenediaminetetraacetic acid), polyphosphates, histidine, tyrosine, and cysteine are the most commonly used chelating agents. Na2-EDTA is the most common chelating agent. EDTA may remove some metal ion components of the cell wall, such as Ca2+, Mg2+, and Zn2+, and may cause cell-wall disintegration. Citric acid is metabolizable by some bacteria. Chelating agents are included in media in low concentrations (e.g., 1 mM).
Growth factors stimulate the growth and synthesis of some metabolites. Vitamins, hormones, and amino acids are major growth factors. Vitamins usually function as coenzymes. Some commonly required vitamins are thiamine (B1), riboflavin (B2), pyridoxine (B6), biotin, cyanocobalamin (B12), folic acid, lipoic acid, p-amino benzoic acid, and vitamin K. Vitamins are required at a concentration range of 10–6 to 10–12 M. Depending on the organism, some or all of the amino acids may need to be supplied externally in concentrations from 10–6 to 10–13 M. Some fatty acids, such as oleic acid and sterols, are also needed in small quantities by some organisms. Higher forms of life, such as animal and plant cells, require hormones to regulate their metabolism. Insulin is a common hormone for animal cells, and auxin and cytokinins are plant-growth hormones.
2.3.3. Growth Media
Two major types of growth media are defined and complex media. Defined media contain specific amounts of pure chemical compounds with known chemical compositions. A medium containing glucose, (NH4)2SO4, KH2PO4, and MgCl2 is a defined medium. Complex media contain natural compounds whose chemical composition is not exactly known. A medium containing yeast extracts, peptone, molasses, or CSL is a complex medium. A complex medium usually can provide the necessary growth factors, vitamins, hormones, and trace elements, often resulting in higher cell yields compared to the defined medium. Often, complex media are less expensive than defined media. The primary advantage of defined media is that the results are more reproducible and the operator has better control of the fermentation. Further, recovery and purification of a product is often easier and cheaper in defined media. Table 2.10 summarizes typical defined and complex media.
2.4. Summary
Microbes can grow over an immense range of conditions: temperatures above boiling and below freezing, high salt concentrations, high pressures (>1000 atm), and at low and high pH values (about 1 to 10). Cells that must use oxygen are known as aerobic. Cells that find oxygen toxic are anaerobic. Cells that can adapt to growth either with or without oxygen are facultative.
The two major groups of cells are procaryotic and eucaryotic. Eucaryotic cells are more complex. The major differences between procaryotic and eucaryotic cells are the absence (procaryotes) or presence (eucaryotes) of a membrane around the chromosomal or genetic material and cellular organelles.
The procaryotes can be divided into two major groups: the eubacteria and archaebacteria. The archaebacteria are a group of ancient organisms; subdivisions include methanogens (methane producing), halobacteria (live in high-salt environments), and thermoacidophiles (grow best under conditions of high temperature and high acidity). Most eubacteria can be separated into gram-positive and gram-negative cells. Gram-positive cells have an inner membrane and strong cell wall. Gram-negative cells have an inner membrane and an outer membrane. The outer membrane is supported by peptidoglycan and the cell envelope is less rigid than in gram-positive cells. The cyanobacteria (blue-green algae) are photosynthetic procaryotes classified as a subdivision of the eubacteria.
The eucaryotes contain both single-celled organisms and multicellular systems. The fungi and yeasts, the algae, and the protozoa are all examples of single-celled eucaryotes. Plants and animals are multicellular eucaryotes.
Viruses are replicating particles that are obligate parasites. Some viruses use DNA to store genetic information, while others use RNA. Viruses that infect bacteria are called bacteriophages or phages.
All cells contain the following macromolecules: protein, RNA, and DNA. Other essential components of the cell are lipids and carbohydrates. Proteins are polymers of amino acids; typically, 20 different amino acids are used. Each amino acid has a distinctive side group. The sequence of amino acids determines the primary structure of the protein. Interactions among the side groups of the amino acids (hydrogen bonding, disulfide bonds, regions of hydrophobicity or hydrophilicity) determine the secondary and tertiary structure of the molecule. If separate polypeptide chains associate to form the final structure, then we speak of quaternary structure. The three-dimensional shape of a protein is critical to its function.
DNA and RNA are polymers of nucleotides. DNA contains the cell’s genetic information that encodes the cell’s proteins (coding DNA). RNA is involved in transcribing and translating that information into real proteins (coding RNA). Messenger RNA transcribes the code; transfer RNA is an adapter molecule that transports a specific amino acid to the reaction site for protein synthesis; and ribosomal RNAs are essential components of ribosomes, which are the structures responsible for protein synthesis. Both DNA and RNA also have noncoding functions. Non-coding DNA is transcribed into functional noncoding RNA molecules (e.g., transfer RNAs, ribosomal RNAs, and regulatory RNAs) and also functions in various aspects of transcriptional and translational regulation. Noncoding RNA functions to regulate gene expression at the transcriptional and posttranscriptional level.
In addition to their role as monomers for DNA and RNA synthesis, nucleotides play important roles in cellular energetics. The high-energy phosphate bonds in ATP can store energy. The hydrolysis of ATP, when coupled to otherwise energetically unfavorable reactions, can drive the reaction toward completion. NAD and NAPH are important carriers of reducing power.
Carbohydrates consist of sugars, and the polymerized products of sugars are called polysaccharides. Sugars represent convenient molecules for the rapid oxidation and release of energy. Polysaccharides play an important structural role (as in cellulose) or can be used as a cellular reserve of carbon and energy (as in starch). Complex sugars or glycans are frequently conjugated to other biomolecules (lipids, proteins) in a process called glycosylation. As attachments to other biomolecules, glycans play important structural, functional, and regulatory roles.
Lipids and related compounds are critical in the construction of cellular membranes. Some fats also form reserve sources. A number of growth factors or hormones involve lipid materials. Phospholipids are the primary components of biological membranes.
The maintenance of cellular integrity requires the selective uptake of nutrients. One class of nutrients is the macronutrients, and these are used in large amounts. The micronutrients and trace nutrients are used in low concentrations; some of these compounds become toxic if present at too high a level.
In a defined medium, all components added to the medium are identifiable chemical species. In a complex medium, one or more components are not chemically defined (e.g., yeast extract).
Suggestions for Further Reading
ALBERTS, B., D. BRAY, K. HOPKIN, A. JOHNSON, J. LEWIS, M. RUFF, K. ROBERTS, AND P. WALTER, Essential Cell Biology, 4th ed., Garland Science, New York, 2013.
HARDIN, J., G. BERTONI, AND L. J. KLEINSMITH, Becker’s World of the Cell, 9th ed., Pearson, Boston, 2015.
MADIGAN, M. T., J. M. MARTINKO, K. S. BENDER, D. H. BUCKLEY, AND D. A. STAHL, Brock Biology of Microorganisms, 14th ed., Pearson, Boston, 2014.
MATHEWS, C. K., K. E. VAN HOLDE, D. R. APPLING, AND S. J. ANTHONY-CAHILL, Biochemistry, 4th ed., Pearson, Toronto, Canada, 2013.
PACE, N. R., Microbial Ecology and Diversity, Am. Soc. Microbiol. News 65: 328–333, 1999.
Questions
2.1. Briefly compare procaryotes with eucaryotes in terms of internal structure and functions.
2.2. Briefly explain the following terms:
a. Heterotrophs
b. Autotrophs
c. Thermophiles
d. Acidophiles
2.3. What are the major differences between eubacteria and archaebacteria?
2.4. Give characteristic dimensions for each of these organisms:
a. E. coli
b. Yeast (S. cerevisiae)
c. Liver cell (hepatocyte)
d. Plant cell
2.5. What are the differences in cell envelope structure between gram-negative and gram-positive bacteria? How might these differences become important if you wish to genetically engineer bacteria to excrete proteins into the extracellular fluid?
2.6. Briefly explain the functions of the following organelles present in eucaryotes:
a. Mitochondria
b. Endoplasmic reticulum
c. Golgi bodies
d. Lysosomes
e. Chloroplasts
2.7. What are the major classes of fungi? Cite the differences among these classes briefly.
2.8. Briefly describe distinct features of actinomycetes and their important products.
2.9. Briefly compare protozoa with algae in terms of their cellular structures and functions.
2.10. Name some industrially important microalgae with their industrial use.
2.11. Cite five major biological functions of proteins.
2.12. Briefly describe the primary, secondary, tertiary, and quaternary structure of proteins. What could happen if tyrosine is substituted for cysteine in the active site? What might happen if the substitution occurred elsewhere?
2.13. Briefly explain the major groups of antibodies and their functions.
2.14. Briefly describe the structure of the following carbohydrates:
a. Sucrose
b. Maltose
c. Lactose
d. Amylose
e. Amylopectin
2.15. What are differences between lipids, fats, and steroids? Why are steroids important to cells?
2.16. Compare structure and functions of DNA and RNA. Cite at least four differences.
2.17. Briefly explain the three major types of RNA and their functions.
2.18. Briefly explain the following terms:
a. Replication
b. Transcription
c. Translation
d. Plasmids
2.19. What are the macronutrients, micronutrients, and trace elements? Provide some examples.
2.20. What are the typical major sources of carbon, nitrogen, and phosphorus in industrial fermentations for antibiotics?
2.21. Explain the functions of the following trace elements in microbial metabolism: Fe, Zn, Cu, Co, Ni, Mn, vitamins.
2.22. What are chelating agents? Explain their function with an example.
2.23. Compare the advantages and disadvantages of chemically defined and complex media.
2.24. You are asked to develop a medium for production of an antibiotic. The antibiotic is to be made in large amounts (ten 100,000 l fermenters) and is relatively inexpensive. The host cell is a soil isolate of a fungal species, and the nutritional requirements for rapid growth are uncertain. Will you try to develop a defined or complex medium? Why?
2.25. You wish to produce a high-value protein using recombinant DNA technology. Would you try to develop a chemically defined medium or a complex medium? Why?