One of the most popular and well-known clustering methods is K-Means clustering. It's conceptually simple. It's also easy to implement and is computationally cheap. We can get decent results quickly for many different datasets.
On the downside, it sometimes gets stuck in local optima and misses a better solution.
Generally, K-Means clustering performs best when groups in the data are spatially distinct and are grouped into separate circles. If the clusters are all mixed, this won't be able to distinguish them. This means that if the natural groups in the data overlap, the clusters that K-Means generates will not properly distinguish the natural groups in the data.
For this recipe, we'll need the same dependencies in our project.clj file that we used in the Loading CSV and ARFF files into Weka recipe.
However, we'll need a slightly different set of imports in our script or REPL:
(import [weka.core EuclideanDistance]
[weka.clusterers SimpleKMeans])For data, we'll use the Iris dataset, which is often used for learning about and testing clustering algorithms. You can download this dataset from the Weka wiki at http://weka.wikispaces.com/Datasets or from http://www.ericrochester.com/clj-data-analysis/UCI/iris.arff. We will load it using load-arff, which was covered in Loading CSV and ARFF files into Weka.
For this recipe, we'll first define a function and macro that will greatly facilitate writing wrapper functions around Weka classes and processes. Then we'll use that macro for the first time to wrap the weka.clusters.SimpleKMeans class:
- First, we'll define a function to generate a random seed from the current time. By default, Weka always uses 1 as the random seed. This function will allow us to use the current time by specifying
nilas the seed:(defn random-seed [seed] (if (nil? seed) (.intValue (.getTime (java.util.Date.))) seed)) - Most of the Weka analyses follows the same pattern. We'll take keyword parameters and turn them into a command-line-style string array. We'll create a Weka object and pass it the options array. This is the kind of boilerplate that the Lisp and Lisp macros are particularly good at abstracting away.
The most complicated part of this will involve parsing a sequence of vectors into the wrapper function's parameter list and the options array. Each vector will list the option, a variable name for it, and a default value. Optionally, they may also contain a keyword indicating how the value is converted to an option and under what circumstances it's included. The highlighted comments in the code show some examples of these near the place where they're passed. Here's the function to parse an options vector into the code to pass to
->options:(defn analysis-parameter [parameter] (condp = (count parameter) ;; [option-str variable-name default-value] ;; ["-N" k 2] 3 '[~(first parameter) ~(second parameter)] ;; [option-str variable-name default-value flag] ;; ["-V" verbose false :flag-true] 4 (condp = (last parameter) :flag-true '[(when ~(second parameter) ~(first parameter))] :flag-false '[(when-not ~(second parameter) ~(first parameter))] :not-nil '[(when-not (nil? ~(second parameter)) [~(first parameter) ~(second parameter)])] :seq (let [name (second parameter)] (apply concat (map-indexed (fn [i flag] '[~flag (nth ~name ~i)]) (first parameter)))) '[~(first parameter) (~(last parameter) ~(second parameter))]) ;; [option-str variable-name default-value flag ;; option] ;; ["-B" distance-of :node-length :flag-equal ;; :branch-length] 5 (condp = (nth parameter 3) :flag-equal '[(when (= ~(second parameter) ~(last parameter)) ~(first parameter))] :predicate '[(when ~(last parameter) [~(first parameter) ~(second parameter)])]))) - Now we can create a macro that takes a name for the wrapper function, a class and a method for it, and a sequence of parameter vectors. It then defines the analysis as a function:
(defmacro defanalysis [a-name a-class a-method parameters] '(defn ~a-name [dataset# & ;; The variable-names and default-values are ;; used here to build the function's ;; parameter list. {:keys ~(mapv second parameters) :or ~(into {} (map #(vector (second %) (nth % 2)) parameters))}] ;; The options, flags, and predicats are used to ;; construct the options list. (let [options# (->options ~@(mapcat analysis-parameter parameters))] ;; The algorithm's class and invocation ;; function are used here to actually ;; perform the processing. (doto (new ~a-class) (.setOptions options#) (. ~a-method dataset#))))) - Now we can define a wrapper for K-Means clustering (as well as the other algorithms we'll introduce later in the chapter) very quickly. This also makes clear how the macro has helped us. It's allowed us to DRY-up (Don't Repeat Yourself) the options list. Now we can clearly see what options an algorithm takes and how it uses them:
(defanalysis k-means SimpleKMeans buildClusterer [["-N" k 2] ["-I" max-iterations 100] ["-V" verbose false :flag-true] ["-S" seed 1 random-seed] ["-A" distance EuclideanDistance .getName]])
- We can now call this wrapper function and get the results. We'll first load the dataset and then filter it into a new dataset that only includes the columns related to the petal size. Our clustering will be based upon those attributes:
user=> (def iris (load-arff "data/UCI/iris.arff")) user=> (def iris-petal (filter-attributes iris [:sepallength :sepalwidth :class])) user=> (def km (k-means iris-petal :k 3)) user=> km #<SimpleKMeans kMeans ====== Number of iterations: 8 Within cluster sum of squared errors: 1.7050986081225123 …
There are several interesting things to talk about in this recipe.
As far as clustering algorithms (or as far as any algorithms) are concerned, K-Means is simple. To group the input data into K clusters, we initially pick K random points in the data's domain space. Now follow the steps listed here:
- Assign each of the data points to the nearest cluster
- Move the cluster's position to the centroid (or mean position) of the data assigned to that cluster
- Repeat
We keep following these steps until either we've performed a maximum number of iterations or until the clusters are stable, that is, when the set of data points assigned to each cluster doesn't change. We saw this in the example in this recipe. The maximum number of iterations was set to 100, but the clusters stabilized after eight iterations.
K-Means clustering does have a number of quirks to be aware of. First, it must be used with numeric variables. After all, what would the distance be between two species in the Iris dataset? What's the distance between Virginica and Setosa?
Another factor is that it won't work well if the natural classifications within the data (for example, the species in the Iris dataset) aren't in separate circles. If the data points for each class tend to run into each other, then K-Means won't be able to reliably distinguish between the classifications.
The following graph shows petal dimensions of the items in the Iris dataset and distinguishes each point by species (shape) and classification (color). Generally, the results are good, but I've highlighted half a dozen points that the algorithm put into the wrong category (some green crosses or yellow diamonds):
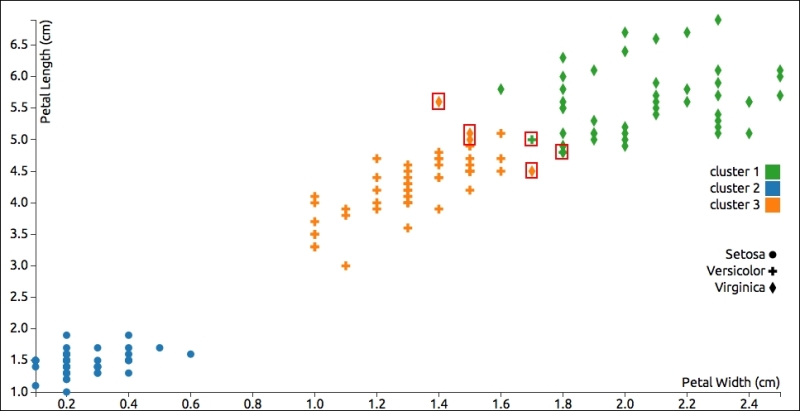
This chart helps us visually verify the results, but the most important part of the algorithm's output is probably the within-cluster sum of squared errors (WCSS). This should be as low as possible for a given number of clusters. In the example, this value is approximately 1.71, which is fine.
Another interesting aspect of this recipe is the use of a macro to create a wrapper function for the cluster analysis. Because Clojure, like other lisps, is written in its own data structures, it's common to write programs to manipulate programs. In fact, this is so common that Clojure provides a stage of compilation dedicated to letting the user manipulate the input program. These meta-programs are called macros. The Clojure compiler reads forms, uses macros to manipulate them, and finally compiles the output of the macros. Macros are a powerful tool that allows users to define their own control structures and other forms far beyond what programmers of other languages have available to them.
We can easily see how the macro is turned into a function using macroexpand-1, like this:
(macroexpand-1 '(defanalysis
k-means SimpleKMeans buildClusterer
[["-N" k 2]
["-I" iterations 100]
["-V" verbose false :flag-true]
["-S" seed 1 random-seed]
["-A" distance EuclideanDistance .getName]]))I cleaned up this output to make it look more like something we'd type. The function that the macro creates is listed here:
(defn k-means
[dataset__1216__auto__ &
{:or {k 2,
max-iterations 100,
verbose false,
seed 1,
distance EuclideanDistance},
:keys [k max-iterations verbose seed distance]}]
(let [options__1217__auto__
(->options "-N" k
"-I" max-iterations
(when verbose "-V")
"-S" (random-seed seed)
"-A" (.getName distance))]
(doto (new SimpleKMeans)
(.setOptions options__1217__auto__)
(.buildClusterer dataset__1216__auto__))))- For information about K-Means clustering, Wikipedia provides a good introduction at http://en.wikipedia.org/wiki/K-means_clustering
- For information about Weka's
SimpleKMeansclass, including an explanation of its options, see http://weka.sourceforge.net/doc.dev/weka/clusterers/SimpleKMeans.html - For more about macros in Clojure, see http://clojure.org/macros. Also, I've written a longer tutorial on macros, which is available at http://writingcoding.blogspot.com/2008/07/stemming-part-8-macros.html
