Another common way to cluster data is the hierarchical way. This involves either splitting the dataset down to pairs (divisive or top-down) or building the clusters up by pairing the data or clusters that are closest to each other (agglomerative or bottom-up).
Weka has a class HierarchicalClusterer to perform agglomerative hierarchical clustering. We'll use the defanalysis macro that we created in the Discovering groups of data using K-Means clustering recipe to create a wrapper function for this analysis as well.
We'll use the same project.clj dependencies that we did in the Loading CSV and ARFF files into Weka recipe, and this set of imports:
(import [weka.core EuclideanDistance]
[weka.clusterers HierarchicalClusterer])
(require '[clojure.string :as str])Because hierarchical clustering can be memory intensive, we'll use the Iris dataset, which is fairly small. The easiest way to get this dataset is to download it from http://www.ericrochester.com/clj-data-analysis/data/UCI/iris.arff. You can also download it, and other datasets, in a JAR file from http://weka.wikispaces.com/Datasets. I loaded it using the load-arff function from the Loading CSV and ARFF files into Weka recipe:
(def iris (load-arff "data/UCI/iris.arff"))
We'll also use the defanalysis macro that we defined in the Discovering groups of data using K-Means clustering recipe.
Now we see some return on having defined the defanalysis macro. We can create a wrapper function for Weka's HierarchicalClusterer in just a few lines:
- We define the wrapper function like this:
(defanalysis hierarchical HierarchicalClusterer buildClusterer [["-A" distance EuclideanDistance .getName] ["-L" link-type :centroid #(str/upper-case (str/replace (name %) - \_))] ["-N" k nil :not-nil] ["-D" verbose false :flag-true] ["-B" distance-of :node-length :flag-equal :branch-length] ["-P" print-newick false :flag-true]]) - Using this, we can filter the petal dimensions fields and perform the analysis:
(def iris-petal (filter-attributes iris [:sepallength :sepalwidth :class])) - Now we can use this data to train a new classifier:
(def hc (hierarchical iris-petal :k 3 :print-newick true))
- To see which cluster an instance falls in, we use
clusterInstance, and we can check the same index in the full dataset to see all of the attributes for that instance:user=> (.clusterInstance hc (.get iris-petal 2)) 0 user=> (.get iris 2) #<DenseInstance 4.7,3.2,1.3,0.2,Iris-setosa>
Hierarchical clustering usually works in a bottom-up manner. Its process is fairly simple:
- Identify the two data points or clusters that are closest to each other
- Group them into a new cluster positioned at the centroid of the pair
- In the population, replace the pair with the new group
- Repeat until we're left with only the number of clusters we expect (the "-N" option mentioned previously)
If we look at a graph of the results, we can see that the results are very similar to what K-Means clustering produced. Actually, there's only one data point that was classified differently, which I've highlighted. In this case, hierarchical clustering grouped that data point incorrectly, possibly pairing it with one of the points to its right:
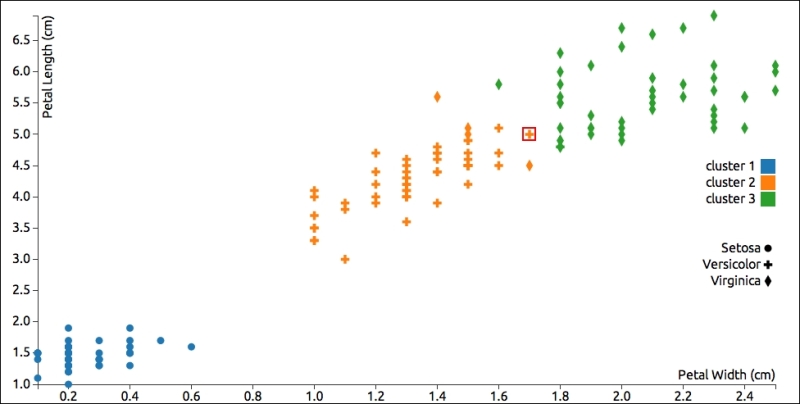
Like K-Means clustering, hierarchical clustering is based on a distance measurement, so it will also have trouble correctly classifying data points along the margins between two groups with close, non-distinct data points.
- The documentation for the
HierarchicalClustererat http://weka.sourceforge.net/doc.dev/weka/clusterers/HierarchicalClusterer.html has more information about the options available for this algorithm - Wikipedia has a good introduction to hierarchical clustering at http://en.wikipedia.org/wiki/Hierarchical_clustering
- David Blei has a set of slides that provides another excellent summary of this algorithm, at http://www.cs.princeton.edu/courses/archive/spr08/cos424/slides/clustering-2.pdf
- The book, Introduction to Information Retrieval, also has a more in-depth look at hierarchical clustering. You can find it online at http://nlp.stanford.edu/IR-book/html/htmledition/hierarchical-clustering-1.html
