One way to classify documents is to follow a hierarchical tree of rules, finally placing an instance into a bucket. This is essentially what decision trees do. Although they can work with any type of data, they are especially helpful in classifying nominal variables (discrete categories of data such as the species attribute of the Iris dataset), where statistics designed for working with numerical data—such as K-Means clustering—doesn't work as well.
Decision trees have another handy feature. Unlike many types of data mining where the analysis is somewhat of a black box, decision trees are very intelligible. We can easily examine them and readily tell how and why they classify our data the way they do.
In this recipe, we'll look at a dataset of mushrooms and create a decision tree to tell us whether a mushroom instance is edible or poisonous.
First, we'll need to use the dependencies that we specified in the project.clj file in the Loading CSV and ARFF files into Weka recipe.
We'll also need these imports in our script or REPL:
(import [weka.classifiers.trees J48]) (require '[clojure.java.io :as io])
For data, we'll use one of the UCI datasets that Weka provides. You can download this set from http://www.cs.waikato.ac.nz/ml/weka/datasets.html, or more directly from http://www.ericrochester.com/clj-data-analysis/data/UCI/mushroom.arff. We can load the datafile using the load-arff function from the Loading CSV and ARFF files into Weka recipe.
We'll also use the defanalysis macro from the Discovering groups of data using K-Means clustering in Weka recipe.
As a bonus, if you have Graphviz installed (http://www.graphviz.org/), you can use it to generate a graph of the decision tree. Wikipedia lists other programs that can display DOT or GV files, at http://en.wikipedia.org/wiki/DOT_language#Layout_programs.
We'll build a wrapper for the J48 class. This is Weka's implementation of the C4.5 algorithm for building decision trees:
- First, we create the wrapper function for this algorithm:
(defanalysis j48 J48 buildClassifier [["-U" pruned true :flag-false] ["-C" confidence 0.25] ["-M" min-instances 2] ["-R" reduced-error false :flag-true] ["-N" folds 3 :predicate reduced-error] ["-B" binary-only false :flag-true] ["-S" subtree-raising true :flag-false] ["-L" clean true :flag-false] ["-A" smoothing true :flag-true] ["-J" mdl-correction true :flag-false] ["-Q" seed 1 random-seed]])
- We can use this function to create a decision tree of the mushroom data, but before that, we have to load the file and tell it which field contains the classification for each one. In this case, it's the last field that tells whether the mushroom is poisonous or edible:
(def shrooms (doto (load-arff "data/UCI/mushroom.arff") (.setClassIndex 22))) (def d-tree (j48 shrooms :pruned true)) - The decision tree outputs Graphviz dot data, so we can write the data to a file and generate an image from that:
(with-open [w (io/writer "decision-tree.gv")] (.write w (.graph d-tree)))
- Now, from the command line, process
decision-tree.gvwith dot. If you're using another program to process the Graphviz file, substitute that here:$ dot -O -Tpng decision-tree.gv
The following graph is created by Graphviz:
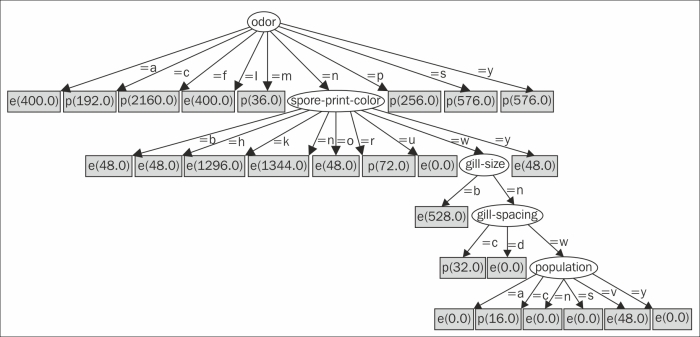
The decision tree starts from the root looking at the odor attribute. Instances that have a value of a (almond) go to the leftmost node, which is marked e (edible). Instances that have a value of c (creosote) go to the next node, which is marked p (poisonous).
However, instances with an odor value of n (none) go to the oval child. It looks at the spore-print-color attribute. If that value is b (buff), then the instance is edible. Other sequences of attributes and values end in different categories of mushrooms.
- Giorgio Ingargiola, of Temple University, has a tutorial page for a class that breaks down the C4.5 algorithm and the ID3 algorithm it's based on, at http://www.cis.temple.edu/~giorgio/cis587/readings/id3-c45.html
- The overview page for the mushroom dataset at http://archive.ics.uci.edu/ml/datasets/Mushroom has information about the attributes used
