In the last recipe, Combining function calls with reducers, we looked at the ability of reducers to compose multiple sequence processing functions into one function. This saves the effort of creating intermediate data structures.
Another feature of reducers is that they can automatically partition and parallelize the processing of tree-based data structures. This includes Clojure's native vectors and hash maps.
For this recipe, we'll continue the Monte Carlo simulation example that we started in the Partitioning Monte Carlo simulations for better pmap performance recipe. In this case, we'll write a version that uses reducers and see how it performs.
From the Partitioning Monte Carlo simulations for better pmap performance recipe, we'll use the same imports, as well as the rand-point, center-dist, and mc-pi functions. Along with these, we also need to require the reducers and Criterium libraries:
(require '[clojure.core.reducers :as r]) (use 'criterium.core)
Also, if you're using Java 1.6, you'll need the ForkJoin library, which you can get by adding this to your project.clj dependencies:
[org.codehaus.jsr166-mirror/jsr166y ""1.7.0""]
- This version of the Monte Carlo pi approximation algorithm will be structured in a similar manner to how
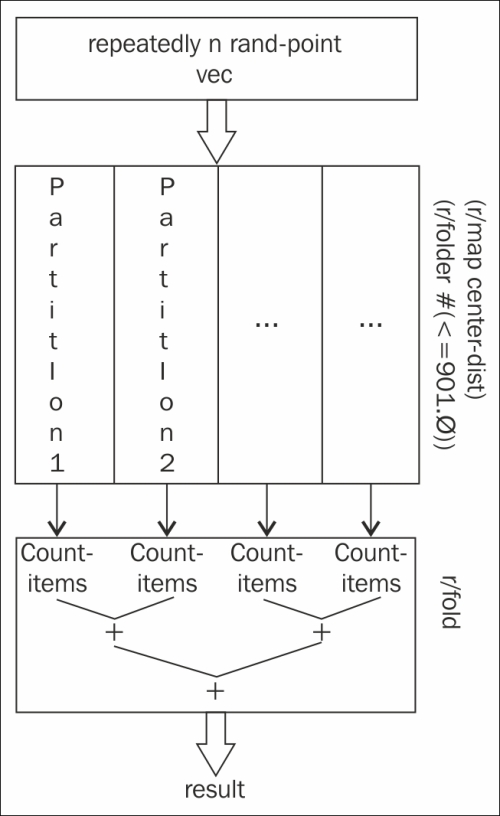
mc-piwas structured in the Partitioning Monte Carlo simulations for better pmap performance recipe. First, we'll define acount-in-circle-rfunction that uses the reducers library to compose the processing and spread it over the available cores:(defn count-items [c _] (inc c)) (defn count-in-circle-r [n] (->> (repeatedly n rand-point) vec (r/map center-dist) (r/filter #(<= % 1.0)) (r/fold + count-items))) (defn mc-pi-r [n] (* 4.0 (/ (count-in-circle-r n) n))) - Now, we can use Criterium to compare the two functions:
user=> (quick-bench (mc-pi 1000000)) WARNING: Final GC required 1.5511109061811519 % of runtime WARNING: Final GC required 1.3658615618441179 % of runtime Evaluation count : 6 in 6 samples of 1 calls. Execution time mean : 983.603832 ms Execution time std-deviation : 16.553276 ms Execution time lower quantile : 964.015999 ms ( 2.5%) Execution time upper quantile : 1.007418 sec (97.5%) Overhead used : 1.875845 ns Found 1 outliers in 6 samples (16.6667 %) low-severe 1 (16.6667 %) Variance from outliers : 13.8889 % Variance is moderately inflated by outliers nil user=> (quick-bench (mc-pi-r 1000000)) WARNING: Final GC required 8.023979507099268 % of runtime Evaluation count : 6 in 6 samples of 1 calls. Execution time mean : 168.998166 ms Execution time std-deviation : 3.615209 ms Execution time lower quantile : 164.074999 ms ( 2.5%) Execution time upper quantile : 173.148749 ms (97.5%) Overhead used : 1.875845 ns nil
Not bad. On eight cores, the version without reducers is almost six times slower. This is more impressive because we made relatively minor changes to the original code, especially when compared to the version of this algorithm that partitioned the input before passing it to pmap, which we also saw in the Partitioning Monte Carlo simulations for better pmap performance recipe.
The reducers library does a couple of things in this recipe. Let's take a look at some lines from count-in-circle-r. Converting the input to a vector was important, because vectors can be parallelized, but generic sequences cannot.
Next, these two lines are combined into one reducer function that doesn't create an extra sequence between the call to r/map and r/filter. This is a small, but important, optimization, especially if we stacked more functions into this stage of the process:
(r/map center-dist)
(r/filter #(<= % 1.0))The bigger optimization is in the line for r/fold. r/reduce always processes serially, but if the input is a tree-based data structure, r/fold will employ a fork-join pattern to parallelize it. This line takes the place of a call to count by incrementing a counter function for every item in the sequence so far:
(r/fold + count-items))))
Graphically, this process looks something similar to this chart:
The reducers library has a lot of promise to automatically parallelize structured operations with a level of control and simplicity that we haven't seen elsewhere.
