1.3 Genome-wide Measurements
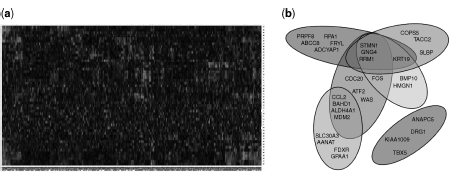
In this section, we present an overview of two complementary forms of data resources (Fig. 1.3), both of which have been utilized by the existing network reconstruction algorithms. The first resource is gene expression data, which is represented as matrix of gene expression levels. The second data resource is a gene set compendium. Each gene set in a compendium stands for a set of genes and the corresponding gene expression levels may or may not be available.
Figure 1.3 Two complementary forms of data accommodated by the existing network reconstruction algorithms. (a) Gene expression data generated from high-throughput platforms, for example, microarray. (b) Gene sets often resulted from explorative analysis of large-scale gene expression data, for example, cluster analysis.
1.3.1 Gene Expression Data
Gene expression data is the most common form of data used in the computational inference of biological networks. It is represented as a matrix of numerical values, where each row corresponds to a gene, each column represents an experiment and each entry in the matrix stands for gene expression level. Gene expression profiling enables the measurement of expression levels of thousands of genes simultaneously and thus allows for a systematic study of biomolecular interaction mechanisms on genome scale. In the experimental procedure for gene expression profiling using microarray, typically a glass slide is spotted with oligonucleotides that correspond to specific gene coding regions. Purified RNA is labeled and hybridized to the slide. After washing, gene expression data is obtained by laser scanning. A wide range of microarray platforms have been developed to accomplish the goal of gene expression profiling. The measurements can be obtained either from conventional hybridization-based microarrays [51–53] or contemporary deep sequencing experiments [54, 55]. Affymetrix GeneChip (www.affymetrix.com), Agilent Microarray (www.genomics.agilent.com), and Illumina BeadArray (www.illumina.com) are representative microarray platforms. Gene-expression data are accessible from several databases, for example, National Center for Biological Technology (NCBI) Gene Expression Omnibus (GEO) [56] and the European Molecular Biology Lab (EMBL) ArrayExpress [57].
1.3.2 Gene Sets
Gene sets are defined as sets of genes sharing biological similarities. Gene sets provide a rich source of data to infer underlying gene regulatory mechanisms as they are indicative of genes participating in the same biological process. It is impractical to collect a large number of samples from high-throughput platforms to accurately reflect the activities of thousands of genes. This poses challenges in gaining deep biological insights from genome-wide gene expression data. Consequently, experimental and computational methods are adopted to reduce the dimension of the space of variables [58]. Such characterizations lead to the discovery of clusters of genes or gene sets, consisting of genes which share similar biological functions. Some of the recent gene set based bioinformatics analyses include gene set enrichment analysis [47–46] and gene set based classification [44, 45]. The major advantage of working with gene sets is their ability to naturally incorporate higher-order interaction patterns. In comparison to gene expression data, gene sets are more robust to noise and facilitate data integration from multiple sources. Computational inference of signaling pathways from gene sets, without assuming the availability of the corresponding gene expression levels, is an emerging area of research [17–20].
