Multidimensional scaling (MDS) can be considered as an alternative to factor analysis when, in addition to the correlation matrices, an arbitrary type of object similarity matrix can be used as input data. MDS is not so much a formal mathematical procedure but rather a method of efficiently placing objects, thus keeping an appropriate distance between them in a new feature space. The dimension of the new space in MDS is always substantially less than the original space. The data that's used for analysis by MDS is often obtained from the matrix of pairwise comparisons of objects. The main MDS algorithm's goal is to restore the unknown dimension,  , of the analyzed feature space and assign coordinates to each object in such a way that the calculated pairwise Euclidean distances between the objects coincide as much as possible with the specified pairwise comparison matrix. We are talking about restoring the coordinates of the new reduced feature space with the accuracy of orthogonal transformation, ensuring the pairwise distances between the objects do not change.
, of the analyzed feature space and assign coordinates to each object in such a way that the calculated pairwise Euclidean distances between the objects coincide as much as possible with the specified pairwise comparison matrix. We are talking about restoring the coordinates of the new reduced feature space with the accuracy of orthogonal transformation, ensuring the pairwise distances between the objects do not change.
Thus, the aim of multidimensional scaling methods can also be formulated in order to display the configuration information of the original multidimensional data that's given by the pairwise comparison matrix. This is provided as a configuration of points in the corresponding space of lower dimension.
Classical MDS assumes that the unknown coordinate matrix, 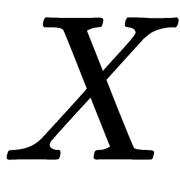 , can be expressed by eigenvalue decomposition,
, can be expressed by eigenvalue decomposition, 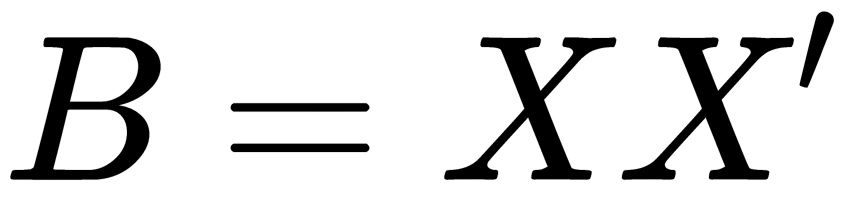 .
.  can be computed from the proximity matrix
can be computed from the proximity matrix 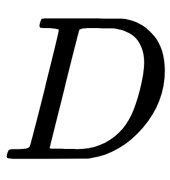 (a matrix with distances between samples) by using double centering. The general MDS algorithm follows these steps:
(a matrix with distances between samples) by using double centering. The general MDS algorithm follows these steps:
- Computes the squared proximity matrix,
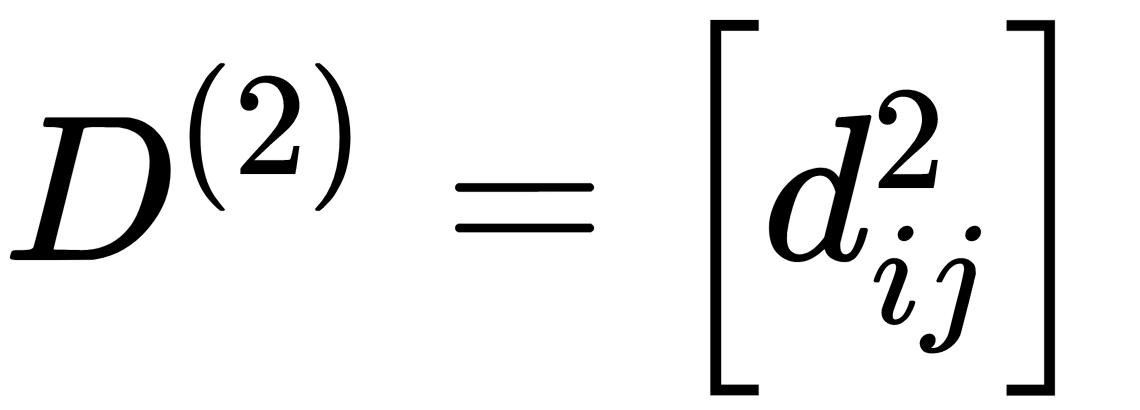 .
. - Applies double centering,
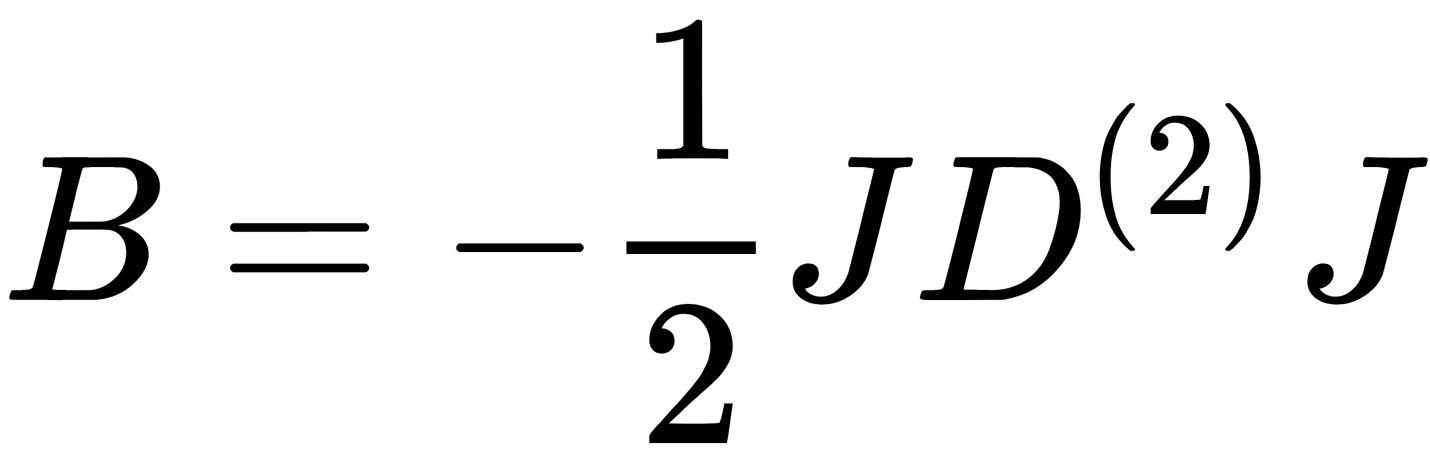 , using the centering matrix,
, using the centering matrix, 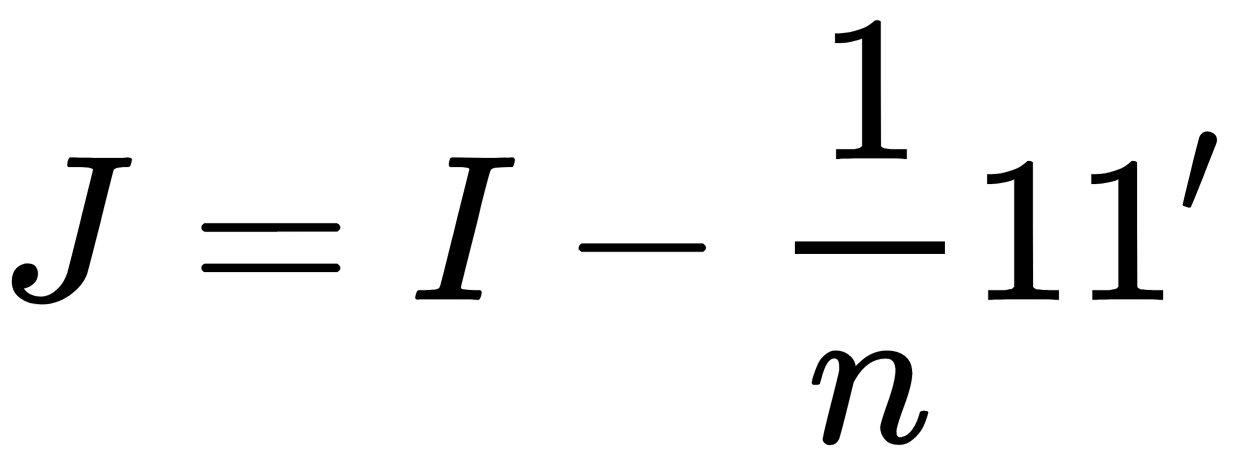 , where
, where 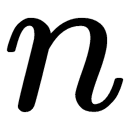 is the number of objects.
is the number of objects. - Determines the
 largest eigenvalues,
largest eigenvalues, 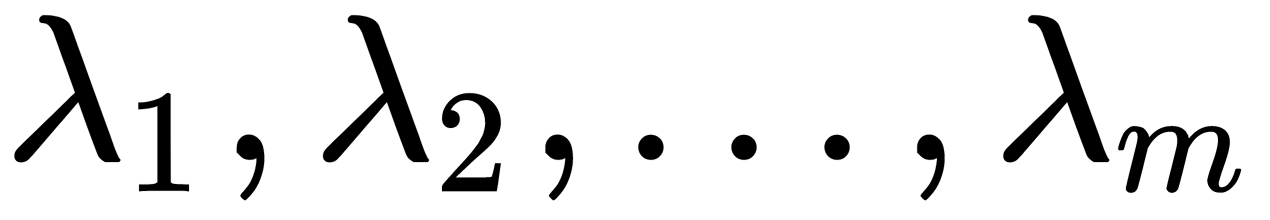 , and the corresponding eigenvectors,
, and the corresponding eigenvectors, 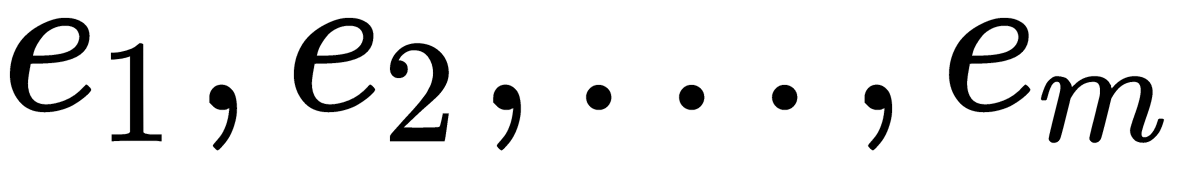 , of
, of  (where
(where 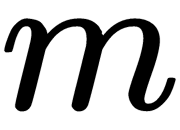 is the number of dimensions desired for the output).
is the number of dimensions desired for the output). - Computes
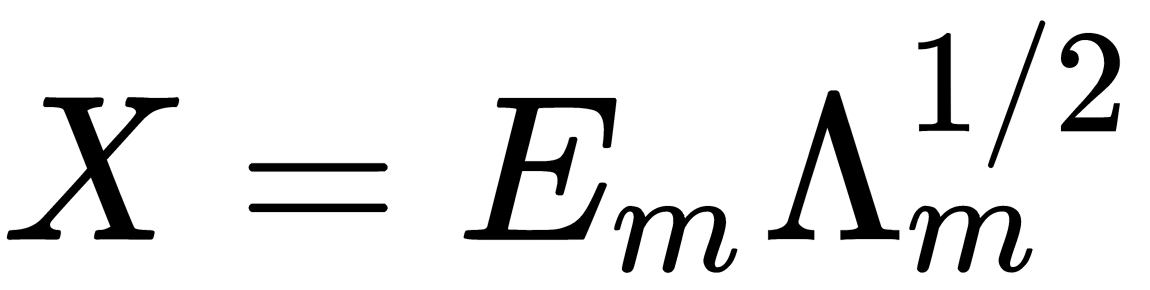 , where
, where 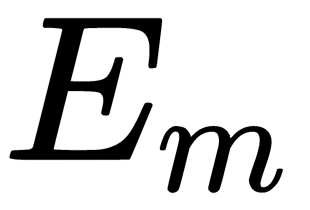 is the matrix of
is the matrix of 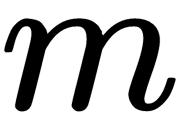 eigenvectors and
eigenvectors and 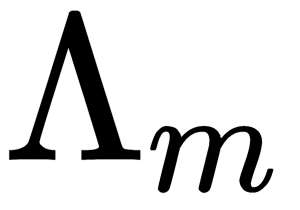 is the diagonal matrix of
is the diagonal matrix of  eigenvalues of
eigenvalues of  .
.
The disadvantage of the multidimensional scaling method is that it does not take into account the distribution of nearby points since it uses Euclidean distances in calculations. If you ever find multidimensional data lying on a curved manifold, the distance between data points can be much more than Euclidean.
Now that we've discussed the linear methods we can use for dimension reduction, let's look at what non-linear methods exist.
