Chapter 1: Predictive analytics and machine learning for medical informatics: A survey of tasks and techniques
Deepti Lamba; William H. Hsu; Majed Alsadhan Department of Computer Science, Kansas State University, Manhattan, KS, United States
Abstract
In this chapter, we survey machine learning and predictive analytics methods for medical informatics. We begin by surveying the current state of practice, key task definitions, and open research problems related to predictive modeling in diagnostic medicine. This follows the traditional supervised, unsupervised, and reinforcement learning taxonomy. Next, we review current research on semisupervised, active, and transfer learning, and on differentiable computing methods such as deep learning. The focus of this chapter is on deep neural networks with common use cases in computational medicine, including self-supervised learning scenarios: these include convolutional neural networks for image analysis, recurrent neural networks for time series, and generative adversarial models for correction of class imbalance in differential diagnosis and anomaly detection. We then continue by assessing salient connections between the current state of machine learning research and data-centric healthcare analytics, focusing specifically on diagnostic imaging and multisensor integration as crucial research topics within predictive analytics. This section includes synthesis experiments on analytics and multisensor data fusion within a diagnostic test bed. Finally, we conclude by relating open problems of machine learning for prediction-based medical informatics surveyed in this chapter to the impact of big data and its associated challenges, trends, and limitations of current work, including privacy and security of sensitive patient data.
Keywords
Predictive analytics; Machine learning; Deep learning; Medical informatics; Health informatics; Prognosis; Diagnosis; Health recommender systems; Integrative medicine
1: Introduction: Predictive analytics for medical informatics
Medical informatics is a broad domain at the intersection of technology and health care which aims to (1) make medical data of patients available to them and to healthcare providers, thus enabling them to make timely medical decisions; and (2) manage this data for educational and research purposes. According to Morris Collen, the first articles on medical informatics appeared in the 1950s (Collen, 1986). However, it was first identified as a new specialty in the 1970s (Hasman et al., 2014).
This section surveys goals, the state of practice, and specific task definitions for machine learning in medical fields and the practice of health care. These sectors produce an enormous amount of data which is highly complex and comes from heterogeneous sources: electronic health records (EHRs) (Thakkar and Davis, 2006), medical equipment and devices, wearable technologies, handwritten notes, lab results, prescriptions, and clinical information. The application of predictive analytics to this data offers potential benefits such as improved standards of care for patients, lower medical costs, and higher resultant patient satisfaction with healthcare providers.
1.1: Overview: Goals of machine learning
Predictive analytics is a branch of data science that applies various techniques including statistical inference, machine learning, data mining, and information visualization toward the ultimate goal of forecasting, modeling, and understanding the future behavior of a system based on historical and/or real-time data. This chapter focuses on machine learning (Samuel, 1959; Jordan and Mitchell, 2015) algorithms for building predictive models. In addition, we will survey applications of machine learning to automation and computer vision, especially image classification, which in some medical domains has achieved accuracy comparable to that of a human expert (Esteva et al., 2017). Sidey-Gibbons and Sidey-Gibbons (2019) provided an introduction to machine learning using a publicly available data set for cancer diagnosis. In recent years, deep learning (LeCun et al., 2015; Goodfellow et al., 2016) has attained technical success and scientific attention in application domains including medicine (Miotto et al., 2018) and health care (Kwak and Hui, 2019). Deep neural networks such as convolutional neural nets (ConvNets or CNNs) have become the predominant state-of-the-art method for analysis of images such as magnetic resonance imaging (MRI) scans, to predict diseases such as Alzheimer’s disease (Liu et al., 2014).
Deep learning models face several challenges in medical domains which hinder their acceptability to the medical community—temporality of data, domain complexity, and lack of interpretability (Miotto et al., 2018). According to Miotto et al. (2018), the most used deep architectures in the health domain, briefly discussed in Section 3, include recurrent neural networks (RNNs) (Schuster and Paliwal, 1997), ConvNets (Lawrence et al., 1997), restricted Boltzmann machines (Nair and Hinton, 2010; Fischer and Igel, 2012), autoencoders (Baldi, 2012; Baxter, 1995), and variations thereof. This chapter focuses on five tasks of medical informatics: differential diagnosis (Sajda, 2006), prediction (Chen and Asch, 2017), therapy recommendation (Gräßer et al., 2017), automation of treatment (Mayer et al., 2008a), and analytics in integrative medicine (Kawanabe et al., 2016).
1.2: Current state of practice
A trend forecast study (Healthcare, 2021) published by the Society of Actuaries indicates a growing usage of predictive analytics for health care. In 2019, 60% of healthcare organizations were already using predictive analytics, and 20% indicated that they would be using the same in the following year. Among those that currently use predictive analytics, 39% reported a decrease in healthcare costs and 42% improvement in patient satisfaction. These statistics demonstrate the interest of organizations in using predictive analytics in medical domain for improving their services.
1.3: Key task definitions
This section provides an overview of machine learning goals in health informatics. The goals of prediction are introduced in Section 1.1.
1.3.1: Diagnosis
Differential diagnosis is defined as the process of differentiating between probability of one disease versus that of other diseases with similar symptoms that could possibly account for illness in a patient. A technical series published by World Health Organization (WHO) in 2016 states that the most important task performed by primary care providers is diagnosis (World Health Organization, 2016). Machine learning tools have been used primarily for disease diagnosis throughout the history of medical informatics. Graber et al. (2005) conducted a study to determine the causes of diagnostic errors and to develop a comprehensive taxonomy for the classification of these errors.
Miller (1994) provided a representative bibliography of the state of the art and history of medical diagnostic decision support systems (MDDSS) at the time. These systems can be divided into several subcategories, among which expert systems have been used most often (Shortliffe et al., 1979). Many of the earliest rule-based expert systems (Giarratano and Riley, 1998) were developed for medical diagnosis. Shortliffe (1986) gave insights into the design of expert systems for diagnostic medicine developed during the 1970s and 1980s, including: (1) MYCIN (Shortliffe and Buchanan, 1985; Shortliffe, 2012), which focused on infectious diseases; (2) INTERNIST-1 (Miller et al., 1982); (3) QMR (Miller and Masarie, 1989; Rassinoux et al., 1996); (4) DXplain (Barnett et al., 1987; mghlcs, n.d.; Bartold and Hannigan, 2002), a diagnostic decision support system developed continuously between 1986 and the early 2000s.
The most prominent limitation of expert systems was the acquisition of knowledge (Gaines, 2013) or building a knowledge base, which is both, time-consuming and a complex process that requires access to expert domain knowledge. In addition, updating the knowledge base requires significant human effort. These systems were usually designed to support users with an expert level of medical knowledge. A more recent review of expert systems is presented by Abu-Nasser (2017). Expert systems are still around but their limitations led to advances in rule learning and classification for differential diagnosis. Salman and Abu-Naser (2020) developed a diagnostic system for COVID-19 using medical websites for the knowledge base. COVID-19 is a novel viral disease that has affected millions of people around the world. The system was tested by a group of doctors and they were satisfied with its performance and ease of use. Another expert system for COVID-19 was built by Almadhoun and Abu-Naser (2020) for helping patients determine if they have been infected with COVID-19. The system gives instructions to the user based on the symptoms. The knowledge base was compiled using medical sites such as NHS Trust.
Kononenko (2001) provided an historical overview of ML methods used in medical domain and a discussion about state-of-the-art algorithms: Assistant-R and Assistant-I (Kononenko and Simec, 1995), lookahead feature construction (Ragavan and Rendell, 1993), naïve Bayesian classifier (Rish et al., 2001), seminaïve Bayesian classifier (Kononenko, 1991), k-nearest neighbors (k-NN) (Dudani, 1976), and back propagation with weight elimination (Weigend et al., 1991). The paper’s experimental findings show that most classifiers have a comparable performance which makes model explainability a deciding factor behind the choice of classifier.
1.3.2: Predictive analytics
Prognosis is defined as a forecast of the probable course and/or outcome of a disease. It is an important task in clinical patient management. Cruz and Wishart (2006) outlined the focal predictive tasks of prognosis/predictions for cancer. Based on these predictive tasks, the general definition of the prognosis task comprises these variants: (1) prediction of disease susceptibility (or likelihood of developing any disease prior to the actual occurrence of the disease); (2) prediction of disease recurrence (or predicting the likelihood of redeveloping the disease after its resolution); and (3) prediction of survivability (or predicting an outcome after the diagnosis of the disease in terms of life expectancy, survivability, disease progression, etc.).
Ohno-Machado (2001) defined prognosis as “an estimate of cure, complication, recurrence of disease, level of function, length of stay in healthcare facilities, or survival for a patient.” The author focused on techniques that are used to model prognosis—especially the survival analysis methods. A detailed discussion of survival analysis methods is beyond the scope of this chapter. We refer the interested readers to the book (Cantor et al., 2003) and a review of survival analysis techniques (Prinja et al., 2010). Prognostic tasks are categorized as (1) prediction for a single point in time and (2) time-related predictions. Methods used to build prognostic models include Cox proportional hazards (Cox and Oakes, 1984), logistic regression (LR) (Kleinbaum et al., 2002), and neural networks (Hassoun et al., 1995).
Mendez-Tellez and Dorman (2005) published an article that states that intensive care units (ICUs) have increased the critical care being provided to injured or critically ill patients. However, the costs for the ICU treatments are very high, which has given rise to prediction models, which are classified as disease-specific or generic models. These systems work by employing a scoring system that assigns points according to illness severity and then generate a probability estimate as an outcome of the model. We do not discuss scoring systems in this chapter. We refer the interested reader to a compendium of scoring systems for outcome distributions (Rapsang and Shyam, 2014). A few of the outcome prediction models used for intensive care predictions include Mortality Probability Model II (Lemeshow et al., 1993), Simplified Acute Physiology Score (SAPS) II (Le Gall et al., 1993), Acute Physiology and Chronic Health Evaluation (APACHE) II (Knaus et al., 1985), and APACHE III (Knaus et al., 1991). These systems build LR models to predict hospital mortality by using a set of clinical and physiology variables.
Another important application of learning is cancer prognosis and prediction. Early diagnosis and prognosis of any life-threatening disease, especially cancer, presents crucial real-time requirements and poses research challenges. Machine learning is being used to build classification models for categorization of cases by risk level. This is essential for clinical management of cancer patients. Kourou et al. (2015) reviewed methods that have been used to model the progression of cancer. The methods used for this task include artificial neural networks (ANNs) (Hassoun et al., 1995) and decision trees (Brodley and Utgoff, 1995), which have been used for three decades for cancer detection. The authors also noticed a growing trend of using methods such as support vector machines (SVMs) (Suykens and Vandewalle, 1999; Vapnik, 2013) and Bayesian networks (BN) (Friedman et al., 1997) for cancer prediction and prognosis.
1.3.3: Therapy recommendation
A classic example of a machine learning application is a recommender system (Portugal et al., 2018; Melville and Sindhwani, 2010). Recommender systems are widely used to recommend items, services, merchandise, and users to each other based on similarity. However, the use of recommender systems in health and medical domain has not been widespread. The earliest article on recommender system in health is from the year 2007 and by 2016 only 17 articles were found for the query “recommender system health” in web of science (Valdez et al., 2016).
Valdez et al. (2016) argued that the lack of popularity of recommender systems in the medical domain is due to several reasons: (1) the benchmarking criteria in medical scenarios, (2) domain complexity, (3) the different end-user groups. The end users or target users for recommender systems can be patients, medical professionals, or people who are healthy. Recommender systems can be designed to recommend therapies, sports or physical activities, medication, diagnosis, or even food or other nutritional information. This chapter also outlines major challenges faced by recommender systems in the medical domain. Challenges include a lack of clear task definition for recommender systems in the health domain. The definition depends on the target user and the item being recommended.
Wiesner and Pfeifer (2014) proposed a health recommender system (HRS) that recommend relevant medical information to the patient by using the graphical user interface of the personal health record (PHR) (Tang et al., 2006). The HRS uses the PHR to build a user profile and the authors argued that collaborative filtering is an appropriate approach for building such a system.
Gräßer et al. (2017) proposed two methods for recommending therapies for patients suffering from psoriasis: a collaborative recommender and a hybrid demographic-recommender. The two are compared and combined to form an ensemble of recommender systems in order to combat drawbacks of the individual systems. The data for the experiments were acquired from University Hospital Dresden. Collaborative filtering (Sarwar et al., 2001; Su and Khoshgoftaar, 2009) is applied, where therapies are items and therapy responses are treated as user preferences.
Stark et al. (2019) presented a systematic literature review on recommender systems in medicine that covers existing systems and compares them on the basis of various features. Some interesting finds from the review include the following observations: (1) most studies attempt to develop the general-purpose recommender systems (i.e., one system for all diseases); (2) disease-specific systems focus on drug recommendation for diabetes. The review points to several future research directions that include building a recommender system for recommending dosage of medicine and finding highly scalable solutions. Recommender systems can be used to suggest drugs for treatment. A popular commercial solution is IBM’s AI machine Watson Health (IBM Watson AI Healthcare Solutions, 2021), which is used by healthcare providers and researchers to make suitable decisions about providing treatment to patients based on insights from the system.
1.3.4: Automation of treatment
In surgical area, research focus has been on automating tasks such as surgical suturing, implantation, and biopsy procedures. Taylor et al. (2016) presented a broad overview of medical robot systems within the context of computer integrated surgery. This article also provides a high-level classification of such systems: (1) surgical CAD/CAM systems and (2) surgical assistants. The former refers to the process of preoperative planning involving the analysis of medical images and other patient information to produce a model of the patient. This article presents examples of both kinds of robotic systems.
Mayer et al. (2008b) developed an experimental system for automating recurring tasks in minimal invasive surgery by extending the learning by demonstration paradigm (Schaal, 1997; Atkeson and Schaal, 1997; Argall et al., 2009). The system consists of four robotic arms which can be equipped with minimally invasive instruments or a camera. The benchmark task selected for this work is minimally invasive knot-tying.
Moustris et al. (2011) presented a literature review of commercial medical systems and surgical procedures. This work solely focuses on systems that have been experimentally implemented on real robots. Automation has also been used for simulating treatment plans on virtual surrogates of patients called phantoms (Xu, 2014). The phantoms represent the anatomy of a patient but they are too generic and hence cannot accurately represent individuals. These phantoms are especially used in pediatric oncology to study the effects of radiation treatment and late adverse effects. Virgolin et al. (2020) proposed an approach to build automatic phantoms by combining machine learning with imaging data. The problem of structuring a pediatric phantom is divided into three prediction tasks: (1) prediction of a representative body segmentation, (2) prediction of center of mass of the organ at risk, and (3) prediction of representative segmentation. Machine learning algorithms used for all three prediction tasks are least angle regression (Efron et al., 2004), least absolute shrinkage and selection operator (Tibshirani, 1996), random forests (RFs) (Breiman, 2001), traditional genetic programming (GP-Trad) (Koza, 1994), and genetic programming—gene pool optimal mixing evolutionary algorithm (Virgolin et al., 2017).
1.3.5: Other tasks in integrative medicine
The Consortium of Academic Health Centers for Integrative Medicine (imconsortium, 2020) defines the term integrative medicine as “an approach to the practice of medicine that makes use of the best-available evidence taking into account the whole person (body, mind, and spirit), including all aspects of lifestyle.” There are many definitions for integrative medicine in the literature, but all share the commonalities that reaffirm the importance of focusing on the whole person and lifestyle rather than just physical healing. According to Maizes et al. (2009), integrative medicine gained recognition due to the realization that people spend only a fraction of time on prevention of disease and maintaining good health. The authors presented a data-driven example to promote the importance of integrative medicine—walking every day for 2 h for adults afflicted with diabetes reduces mortality by 39%. It is important to note that integrative medicine is not synonymous with complementary and alternative medicine (CAM) (Snyderman and Weil, 2002). We refer interested readers to Baer (2004), which chronicles the evolution of conventional and integrative medicine in the United States.
CAM refers to medical products and practices that are not part of standard medical care. Ernst (2000) presented examples of techniques used in CAM which include but are not limited to the following: acupuncture, aromatherapy, chiropractic, herbalism, homeopathy, massage, spiritual healing, and traditional Chinese medicine (TCM).
Zhao et al. (2015) presented an overview of machine learning approaches used in TCM. TCM specialists have established four diagnostic methods for TCM: observation, auscultation and olfaction, interrogation, and palpation. This article explains each of the four diagnostic methods and provides a list of machine learning methods used for each task. The most common methods are kNNs and SVM. Other methods include decision trees, Naïve Bayes (NB), and ANNs.
1.4: Open research problems
A recently published editorial by Bakken (2020) highlights five clinical informatics articles that reflect a consequentialist perspective. One of the articles that we discuss here focuses on a methodological concern, that is, predictive model calibration (Vaicenavicius et al., 2019). Predictive models are an important research topic as discussed in Section 1.3.2, but many studies continue to focus on model discrimination rather than calibration. Ghassemi et al. (2020) outlined several promising research directions, specifically highlighting issues of data temporality, model interpretability, and learning appropriate representations. Machine learning models in most of the existing literature have been trained on large amount of historical data and fail to account for temporality of data in the medical domain, where patient symptoms and or treatment procedures change with time. The authors cited Google Flu Trends as an example of the need to update machine learning models to account for this data temporality, as it persistently overestimated flu (Lazer et al., 2014). Another promising research area is model interpretability (Ahmad et al., 2018; Chakraborty et al., 2017). The authors suggested many directions toward the achievement of this goal: (1) model justification to justify the predictive path rather than just explaining a specific prediction; (2) building collaborative systems, where humans and machines work together. A final research topic is representation learning, which can improve predictive performance and account for conditional relationships of interest in the medical domain.
1.4.1: Learning for classification and regression
Classification is the identification of one or more categories or subpopulations to which a new observation belongs, on the basis of a training data set containing observations, or instances. In the data sciences of statistics and machine learning, classification may be supervised (where class labels are known) (Caruana and Niculescu-Mizil, 2006), unsupervised (where they are not and assignment is based on cohesion and similarity among instances) (Ghahramani, 2003), or semisupervised. Dreiseitl and Ohno-Machado (2002) surveyed early work using LR (particularly the binomial logit model) and ANNs (particularly multilayer perceptrons or MLP) for dichotomous classification, also known as binary classification or concept learning, on diagnostic and prognostic tasks from 72 papers in the existing literature. In parallel with this broad study of discriminative approaches to diagnosis and prognosis, Dybowski and Roberts (2005) compiled a comprehensive anthology of probabilistic models primarily for generative classification.
In contrast with these broad surveys, which are included for completeness and historical breadth, application papers tend to focus on specific use cases for classification, such as prediction of mortality. Eftekhar et al. (2005) presented one such paper which addresses the task of predicting head trauma mortality rate based on initial clinical data, and focuses methodologically on LR and MLP, as do Dreiseitl and Ohno-Machado (2002).
Regression is the problem of mapping an input instance to a real-valued scalar or tuple, which in data science is defined as an estimation task. In medical informatics, many predictive applications can be formulated as risk analysis tasks, that is, tasks requiring estimation of syndrome probability, given data from electronic medical records. Typical examples include estimating risk of a particular form of cancer, such as in a study by Ayer et al. (2010), where they use LR and MLP to estimate risk of breast cancer. In some additional use cases, the predictive task requires estimation of a continuous value such as the size (widest diameter) of a cancer mass, rather than a probability of occurrence. Royston and Sauerbrei (2008) presented a methodological introduction to numerical estimation methods for such tasks.
1.4.2: Learning to act: Control and planning
Another general category of tasks falls under the rubric of learning to act, or intelligent control and planning in engineering terminology. This includes the application of machine learning to the overlapping subarea of optimal control, the branch of mathematical optimization that deals with maximizing an objective function such as cost-weighted proximity to a target.
One example of an optimal control task, which was investigated by Vogelzang et al. (2005), is maintaining a patient’s blood glucose level via automatic control of an insulin pump. The functional requirement of the system is to regulate the change in pump rate as a function of past pump rate, target glucose level, and past blood glucose measurements. The Glucose Regulation in Intensive care Patients (GRIP) system developed by Vogelzang et al. (2005) used a fixed weighted optimal control function based on previous clinical studies. Other optimal control tasks include prolonging the onset of drug resistance in treatment applications such as chemotherapy, a task studied by Ledzewicz and Schättler (2006), who formulated a dynamical system for the development of drug resistance over time and applied ordinary differential equation solvers to the task. Such numerical models can also be developed for therapeutic objectives such as minimizing tumor volume as a function of angiogenic inhibitors administered over time, an optimal control task studied by Ledzewiecz et al. (2008).
By formulating parametric models for problems such as maintaining a patient’s healthcare characteristics (e.g., blood glucose level, tumor size) within desired ranges while minimizing the total cost of doing so, optimization methods from industrial engineering and operations research, such as control charts, can be used. For example, Dobi and Zempléni (2019) applied Markov chain models and a variety of control charts to this cost-optimal control task (Dobi and Zempléni, 2019).
Yet another family of intelligent control approaches originates from classical planning, particularly the inverse problem of plan recognition (mapping from observed action sequences to individual plan steps, preconditions, and desired postconditions) and the problem of plan revision, which entails modifying a plan (such as a course of drug therapy) due to an identified complication (such as a toxic episode or other adverse reaction or interaction). Such systems are discussed by Shahar and Musen (1995). Plan revision may be necessitated as a consequence of historical observation (case studies), predictive simulation, or inference using a domain theory.
Finally, enterprise resource planning (ERP) is an integrative planning task of managing business processes (in health care, these include sales and marketing, patient services, provider human resources, specialist referrals, procurement of equipment and materials, treatment, billing, and insurance). van Merode et al. (2004) surveyed ERP requirements and systems for hospitals.
1.4.3: Toward greater autonomy: Active learning and self-supervision
Machine learning depends on availability of a training experience, but this experience needs not come in the form of labeled data, which is expensive to acquire even with copious available resources such as expertise and nonexpert annotator time, whose cost may be reduced by gamification or other means of crowd sourcing to volunteers. In this section, we survey three species of learning without full supervision that help to free machine learning users from some aspects of these data requirements and other experiential requirements. These are
- 1. active learning (Settles, 2009), the problem of developing a learning system that can seek out its own experiences;
- 2. transfer learning (Pan and Yang, 2009), training on a set of experiences for one or more tasks or domains and using the resulting representations to facilitate learning, reasoning, and problem solving in new tasks and domains; and
- 3. self-supervised learning (Ross et al., 2018), the generalized task of learning with unlabeled data and inducing intrinsic labels by discovering relationships between subcomponents of the training input (such as views or parts of an object in computer vision tasks).
Active learning in medical informatics spans a gamut of experiential domains from text to case studies, to controllers and policies. An example of active learning in text is the work of Druck et al. (2009), who applied categorical feature labels to words. This is a typical methodology in biomedical texts, where domain lexicons are organized into syndromic, pharmaceutical, and anatomical hierarchies, among others. Chen et al. (2012) used active learning with labels on two text categorization tasks. The first of these is at the sentence level, on the ASSERTION data set, a clinical healthcare text corpus for the 2010 i2b2/VA natural language processing (NLP) challenge. The second is at the whole-document level, on the NOVA data set, an email corpus with labels corresponding to a generic “religion versus politics” topic classification task. In subsequent work, Chen et al. (2017) applied a conditional random field (CRF)-based active learning system to the corpus of the 2010 i2b2/VA NLP challenge to show how annotation time could be reduced: by using latent Dirichlet allocation for sentential topic modeling (sentence-level clustering) and by bootstrapping the process of set expansion for the named entity recognition (NER) task using active learning. Dligach et al. (2013) also developed an active learning system for the i2b2 task, but focused on document-level phenotyping (prefiltering of ICD-9 codes, CPT codes, laboratory results, medication orders, etc. followed by category labeling). In their work, a document consists of EHRs and all associated data for a given patient, which may be generated at multiple stages of an admission and treatment workflow.
Transfer learning is typically defined as task to task or domain to domain but what constitutes a domain in medical informatics can vary. Wiens et al. (2014) investigated interhospital transfer learning by training on subsets of 132,853 admissions at three different hospitals, among which 1348 positive cases of Clostridium difficile infection were diagnosed, to boost hospital-specific precision and recall as measured holistically using the area under the receiver operating characteristic curve. As with active learning, transfer learning can be based on natural language features, typically at the word, sentence, or document level for medical informatics. For example, Lee et al. (2018) outlined a neural network approach to deidentification in patient notes, which is an instance of NER and crucial for compliance with patient confidentiality laws such as the Health Insurance Portability and Accountability Act in the United States. They applied a long short-term memory (LSTM), a type of deep learning neural network for sequence modeling, to classify named entities that represented protected health information. The transfer learning task, training on a large labeled data set to a smaller one with fewer labels, is another case of interdomain transfer. Yet another NER transfer learning problem for EHRs is defined and studied by Wang et al. (2018), who applied a bidirectional LSTM (Bi-LSTM) on a shared training corpus to create a shared representation (specifically, a word embedding) for text classification that is then fine-tuned for source and target domains by training a CRF model with labeled training data as available, to achieve label-aware NER. The experimental corpus, in this case, is a Chinese-language medical NER corpus (CM-NER) consisting of 1600 anonymized EHRs across four departments: cardiology (500), respiratory (500), neurology (300), and gastroenterology (300). Wang et al. (2018) demonstrated effective interdepartmental NER (domain-to-domain) transfer through experiments on CM-NER. Similar deep learning approaches for NLP are applied by Du et al. (2018), who demonstrated transfer using RNNs from clinical notes on psychological stressors to tweets on Twitter, to detect posts by users at risk of suicide. New architectures for implementing transfer are demonstrated by Peng et al. (2019), who used the deep bidirectional transformers BERT and ELMo to achieve cross-domain transfer among 10 benchmarking data sets from the Biomedical Language Understanding Evaluation (BLUE) compendium.
Self-supervised learning consists of generating labels by means of (1) comparing objects (e.g., clustering), (2) extracting relationships, or (3) designing experiments (especially model selection). The first approach applies similarity or distance metrics over entire instances (unlabeled examples) and has traditionally been similar to unsupervised learning for classification tasks in general, while the second involves using relational patterns and/or probabilistic inference over structured data models to capture new relationships, and the third involves generating multiple candidate models (by random sampling or parameter optimization methods such as identifying support vectors for large margin discriminative classifiers), and then applying model selection.
Hoffmann et al. (2010) took the second approach, introducing LUCHS, a self-supervised, relation-specific system for information extraction (IE) from text. Roller and Stevenson (2014) also used a relation-specific, ontology-aware approach to relation extraction; rather than being based on lexicon expansion, however, it uses the curated Unified Medical Language System, a biomedical knowledge base.
Stewart et al. (2011) presented an application of the third approach to the task of event detection in the domain of social media-based epidemic intelligence, where self-supervised learning consists of tokenizing text corpora, namely ProMED-Mail and WHO outbreak reports, to obtain bag of words (BOW or word vector space) embeddings. An SVM classifier is then trained, to which the authors applied model selection, testing the result against an avian flu text corpus.
As Blendowski et al. (2019) noted, self-supervision in medical imaging applications is a necessity because of the comparatively high cost of supervision for medical images and video versus general computer vision and video. In medical domains, annotation may require specialization in radiology and other medical subdisciplines, whereas generic images and videos may be annotated via microwork systems such as Amazon Mechanical Turk, or even via volunteer crowdsourcing. Blendowski et al. presented a modern, deep learning-based approach to self-supervision, applying convolutional neural networks (CNNs) to capture 3D context features.
2: Background
2.1: Diagnosis
2.1.1: Diagnostic classification and regression tasks
Nadeem et al. (2020) presented a very comprehensive survey on classification of brain tumor. Jha et al. (2019) evaluated 32 supervised learning methods across 17 classification data sets (in domains that include cancer, tumors, and heart and liver diseases) to determine that decision tree-based methods perform better than others on these data sets. Mostafa et al. (2018) used three classification methods (decision trees, ANNs, and NB) to determine the presence of Parkinson’s disease by using features extracted from human voice recordings, reaching a similar conclusion that decision trees performed best on this data set.
Polat and Güneş (2007) presented a binary classification task for categorizing breast cancer as malignant or benign. The authors used least square support vector machine (LS-SVM) (Suykens and Vandewalle, 1999) for classification.
Another machine learning task used for diagnosis is regression. Kayaer et al. (2003) used general regression neural network (GRNN) (Specht et al., 1991) for diagnosing diabetes using the Pima Indian diabetes data set (http://archive.ics.uci.edu/ml). The results show that it performs better than standard MLP and radial basis function (RBF) feedforward neural networks (Broomhead and Lowe, 1988) that have been used by other studies using the same data set. Hannan et al. (2010) have also used GRNN and RBF for heart disease diagnosis. Jeyaraj and Nadar (2019) proposed a regression-based partitioned deep CNN for the classification of oral cancer as malignant or benign. The network obtains accuracy comparable to that of a human expert oncologist.
2.1.2: Diagnostic policy-learning tasks
Yu et al. (2019b) presented the first comprehensive survey of reinforcement learning (RL) applications in health care. The aim of the survey is to provide the research community with an understanding of the foundations, methodologies, existing challenges, and recent applications of RL in healthcare domain. The range of applications vary from dynamic treatment regimes (DTRs) in chronic diseases and critical care, automated clinical diagnosis, to other tasks such as clinical resource allocation and scheduling.
Early RL systems for medical informatics were predominantly designed for medical image processing and analysis (as a specialized application of computer vision). Sahba et al. (2006) developed such a system, based on Q-learning, for ultrasound image analysis, focusing on tasks such as local thresholding, feature extraction, and segmentation of organs (in this case, the prostate gland). In subsequent work, Sahba et al. (2007) extended this Q-learning system for organ segmentation to an adversarial framework that they termed “Opposition-Based Learning.” This framework allowed for more flexible formulation of utility gradients for RL problems such as balancing exploration versus exploitation.
Peng et al. (2018) introduced REFUEL, a deep Q-network (DQN)-based system for reward shaping and feature construction (“rebuilding”) in differential diagnosis of diseases. Such systems are examples of reinforcement-based metalearning and can potentially incorporate aspects of both self-supervision and active learning of representation. DQN has also been used by researchers such as Al and Yun (2019) to learn policies for recognizing anatomical landmarks in X-ray-based computerized tomography (CT) and MRI images. In addition, DQN has recently been applied to learn control policies for clinical decision support tasks such as guiding a healthcare professional or first responder, especially an emergency medical technician, in obtaining ultrasound images as a remote sensing step before administering treatment. Milletari et al. (2019) presented a novel application of this method to Point of Care Ultrasound (POCUS) for scanning the left ventricle of the heart.
Utility functions for deep RL in medical informatics may be tied to anatomical mapping and other automation tasks of internal medicine. Examples of such mapping include the context maps of Tu and Bai (2009), which are based on learning, i.e., parameter estimation, in Markov random fields and CRFs. In this work, the authors trained and applied inference using these models to solve high-level vision tasks such as image segmentation, configuration estimation (orientation), and region labeling of 3D brain images.
A key family of RL applications is that of control policies for medical treatment. Weng et al. (2017) described a deep RL method using a sparse autoencoder for glycemic control in septic patients. While experimental validation of this system was performed using historical data, the RL framework presented can include medical devices and mixed-initiative systems.
Finally, RL can also be applied to interactive differential diagnosis using natural language (i.e., dialogue). Liu et al. (2018) described a dialogue-based system for disease phenotype identification (eliciting observable characteristics or traits of diseases, such as the presentation and development of symptoms, morphology, biochemical or physiological properties, or patient behavior). The dialogue policy formulated here is a Markov decision process (MDP). The RL problem is that of detecting symptoms of any or all four known pediatric diseases by simulating query-based dialogue using an annotated corpus. The authors showed that DQN for dialogue outperforms SVM for supervised classification learning, random dialogue generation, and a rule-based dialogue agent.
2.1.3: Active, transfer, and self-supervised learning
Sánchez et al. (2010) proposed a computer aided diagnosis (CAD) (Castellino, 2005) system for diabetic retinopathy screening using active learning approaches. There are four components that constitute the DR screening process: quality verification, normal anatomy detection, bright lesion detection, and red lesion detection (Niemeijer et al., 2009). The findings from these four components need to be fused in order to generate an outcome for a patient. The outcome is in the form of a likelihood that the patient will be referred to an ophthalmologist. The output from the four components of the DR screening are used to extract some features which are further used to train a kNN classifier. Active learning is applied in the training phase to select an unlabeled sample from the pool of samples and pose a query to the expert in order to acquire a label for the sample. This is an iterative process that only stops when a stopping criterion has been reached. This work used two different query functions: uncertainty sampling (Lewis and Gale, 1994) and query-by-bagging (QBB) sampling (Abe, 1998).
Apostolopoulos and Mpesiana (2020) used a transfer learning approach for the classification of medical images to diagnose COVID-19. This study uses publicly available thoracic X-rays of healthy people as well as patients suffering from COVID-19 to build an automatic diagnostic system. The aim of their work is to evaluate the effectiveness of state-of-the-art pretrained CNN models for the diagnosis of COVID-19. The CNN used for this study include VGG19 (Simonyan and Zisserman, 2015), MobileNetv2 (Sandler et al., 2018), Inception (Szegedy et al., 2015), Xception (Chollet, 2017), and InceptionResNet v2 (Szegedy et al., 2017). The study formulates the task as a multiclass classification problem with three classes: normal people, pneumonia patients, and COVID-19 patients. The study does accomplish its goals of establishing the benefits of transfer learning by using state-of-the-art CNN models.
Bai et al. (2019) used a semisupervised approach for learning features from unlabeled data for the task of cardiac MR image segmentation. This segmentation is important for characterizing the function of the heart. The authors discussed the different angulated planes at which the MR images are acquired. For brevity, we refer the readers to Bai et al. (2019). Specific views of the scans and their labels have been traditionally used to train a network from scratch. The authors used a standard U-Net architecture (Ronneberger et al., 2015) with three variations to it. The results of their work show that by using self-supervised learning even a small data set is able to outperform a standard U-Net that has been trained from scratch.
2.2: Predictive analytics
2.2.1: Prediction by classification and regression
We start by discussing prediction of disease susceptibility: Kim and Kim (2018) tried to predict an individual’s susceptibility to cancer by using genomic data. The authors used kNN for building a multiclass classification model.
Next we move on to prediction of survivability: Choi et al. (2009) proposed a hybrid ANN and BN model to predict 5-year survival rates for breast cancer patients. The model combines the best of both worlds using black box ANN for their higher accuracy and BNs for their explainability.
The survivability of a cancer patient depends on the stage of cancer, which is based on tumor size, location, spread, and other factors. Machine learning models that predict survivability in breast cancer research usually use breast cancer stage as a feature for training the model. Kate and Nadig (2017) referred to such a model as a joint model. Their work used the SEER data set (SEER Incidence Database—SEER Data & Software, 2021), which classifies cancer into four stages: in situ, localized, regional, and distant.
Mobadersany et al. (2018) proposed an approach called Survival Convolutional Neural Network (SCNN), which uses a CNN integrated with Cox survival analysis technique for the task of survivability prediction for patients with brain tumors. The CNN includes a Cox proportional hazards layer that models overall survival. The proposed approach surpasses the prognostic accuracy of human experts.
The final task that we discuss is prediction of recurrence: The task involves correctly predicting the recurrence of disease with a binary outcome. Abreu et al. (2016) presented a literature review to evaluate the performance of machine learning methods for the task of predicting breast cancer recurrence. The review covers literature during the years 2007–14 and find that the key algorithms used for the task include: decision trees, LR, ANN, NB, K-Means, RFs, and kNN. These algorithms are discussed in Section 3. The authors outlined a few challenges based on their review that make this task less popular: (1) lack of publicly available data of reasonable size—most of the studies have used local data sets usually with a small number of patients; (2) data imbalance is not handled in most cases; (3) feature selection was performed manually in most studies with the help of domain experts—there is no agreement on variables that are important for the study of breast cancer recurrence; (4) accuracy as evaluation metric for classification performance which is not appropriate for imbalanced data sets; and (5) lack of model interpretability of machine learning models—these challenges provide a scope for future research in the area. A more recent review was published by Zhu et al. (2020) who covered the usage of deep learning for the task of cancer prognosis. The study also presents similar challenges encountered by deep learning models in the domain as were pointed by Abreu et al. (2016). Some of the challenges listed include: (1) availability of small data sets; (2) handling data imbalance; (3) handling sparse and missing data; (4) handling high-dimensional sequencing data; and (5) need of researchers with expertise in both, machine learning and biomedical domain.
2.2.2: Learning to predict from reinforcements and by supervision
The advent of institution-wide terascale to petascale data mining, considered “big data” as of 2020, has brought machine learning for predictive analytics to the fore in many clinical domains. Shah et al. (2018) presented a commentary piece on the state of the field in data mining for predictive analytics in medical informatics. It cites the CHA2DS2 − V ASc score as an example of a predictive rule regarding the doubling of thromboembolic risk in atrial fibrillation as a function of congestive heart failure, hypertension, age, diabetes, and previous stroke or transient ischemic attack, citing it as a use case of predictive models with consequence for finely balanced treatment decisions, which the authors note are commonplace. Rajkomar et al. (2019) reviewed a broader set of diagnostic and prognostic applications, along with best practices for using machine learning as an augmentative technology for clinicians. Shameer et al. (2018) surveyed the field of artificial intelligence (AI) in medicine even more broadly, discussing the species of machine learning surveyed in this chapter and the actionable products thereof, from classification rules and regression formulas to annotated images and policies from RL.
Many predictive analytics applications involve estimating risk of adverse effects, or detecting imminent adverse outcomes in time for preventative or anticipatory measures. For example, Kendale et al. (2018) developed an LR-based system for predicting hypotension in patients after surgical anesthesia.
Just as supervised learning finds precedent in rule-based expert systems for differential diagnosis, RL finds precedent in both educational technology and early automation for the practice of internal medicine and general surgery. Examples are surveyed in Section 4. One general use case of RL for predictive analytics is given by Khurana et al. (2018), who cast feature engineering for predictive applications as an RL task where the policy is defined in terms of data transformations.
Taking such diverse use cases as a whole, a major consideration in medical predictive analytics, which overlaps with AI, ethics, and society (AIES), as well as with AI safety and security, is how to determine appropriate regulatory standards of clinical benefit. Parikh et al. (2019) discussed meaningful minimum standards of functionality and interoperability, in the context of endpoints (clinical outcomes), appropriate benchmarks, and the importance of associating predictive systems with interventions.
2.2.3: Transfer learning in prediction
Gliomas are a type of tumors that are found in the brain. They are of different types and are usually graded from I to IV with grade IV being the most aggressive type. MRI images are used for grading gliomas into two categories: lower-grade glioma (LGG), which is grade II and III and higher-grade glioma, which is a grade IV. Cabezas et al. (2018) proposed a model using CNN for classifying LGG and HGG by analyzing MRI images. Two CNN architectures were explored for this purpose, namely: AlexNet and GoogLeNet. These two architecture are discussed in Section 3.3.2. The results indicate that with transfer learning and fine-tuning the performance improved for both deep learning architectures (DLAs). Yang et al. (2018) presented a transfer learning approach to segment the gliomas and its subregions and use the results along with other clinical features to predict patient survival. The approach is divided into two main tasks, where the first task focuses on segmentation of glioma using a 3D U-Net (Ronneberger et al., 2015). The second task segments the tumor subregions using a small ensemble net (Kamnitsas et al., 2017). Their work uses a VGG-16 network, which is discussed in Section 3.3.2.
2.3: Therapy recommendation
2.3.1: Supervised therapy recommender systems
Therapy recommendation remains a common use case of supervised learning, beyond diagnosis. General-purpose models for classification such as nearest-neighbor and rule-based classification are often effective, particularly when explainability is desired. Zhang et al. (2013) discussed supervised learning for therapy recommendation in the domain of physical therapy, where class imbalance is a frequent issue, and derive both a modified rule-learning algorithm (ARIPPER) and an application of the selective minority oversampling technique (SMOTE) for this task.
Mental health and wellbeing are major health issues in the world. There is a growing research effort to alleviate the symptoms of depression. Rohani et al. (2020) built a recommender system for the mental health domain. The authors cited the motivation behind such systems stemming from research that suggests that when participants regularly participate in a pleasant activity then it has a beneficial impact on the mental health of the individual (MacPhillamy and Lewinsohn, 1982; Fredrickson, 2000). An effective treatment for depression has been the pleasant event scheduling system (Lewinsohn and Libet, 1972). Wahle et al. (2016) described multiuser and personalized variants of an affective recommender system developed using data from two mobile health applications for both clinically depressed users and nonclinical users. Mood ratings for activities are predicted using trained NB and SVM models. These classification-based predictions enable personalized treatment recommendations using a mobile app.
2.4: Automation of treatment
2.4.1: Classification and regression-based tasks
Learning in the presence of hybrid training data (consisting of continuous and discrete or nominal variables) is a frequent and typical necessity in medical AI applications. Schilling et al. (2016) described the use of Classification and Regression Trees (CART) to identify thresholds for cardiovascular disease. Such hybrid tasks can also be addressed using binary response models such as probit models, but as the authors note, three distinct strengths of CART are that: (1) its hierarchical structure makes it more human comprehensible than linear or LR models; (2) it can capture nonlinearity in response and some multivariate interactions; and (3) it has more expressiveness than regression models.
For some predictive applications, accuracy, precision, and recall are of highest significance and may be considered more important than explainability by diagnosticians. Taylor et al. (2018) presented a task where regression methods, gradient boosting improvements, and committee machines such as bagged RFs and Adaboost are highly effective. In some cases, association rule mining and basic NLP methods suffice: De Silva et al. (2018) described a social media mining task, analytics of online social groups for cancer patients, where the technical objective is to extract information about user demographics in relation to terms indicating patient age, cancer stage, side effects reported, and sentiments.
2.4.2: RL for automation
An essential task in automation of surgical tasks is optimal path planning to avoid collisions between tool and surrounding tissue before resection automation. Baek et al. (2018) aimed to create a global path to cut a tissue during surgery without colliding with the surrounding tissue. The generated path was simulated on APOLLON, which is a Single Incision Laparoscopic Surgery system developed by KAIST Telerobotics and Control Lab (Medical Robot|KAIST Telerobotics and Control Lab, 2021). Their work uses a popular path planning method called probabilistic roadmap (RPM) (Kavraki et al., 1998), which creates a path from a static environment to a desired point and Q-learning (Watkins and Dayan, 1992), an RL technique.
2.4.3: Active learning in automation
In a hospital setting, hand hygiene is one of the most important factors for the prevention of infectious diseases. Monitoring hand hygiene could be vital in reducing any outbreak within an operating room (OR). Kim et al. (2020) proposed a fully augmented automatic hand hygiene monitoring tool for monitoring the anesthesiologists on OR video. The aim is to identify the alcohol-based hand rubbing actions of anesthesiologists in the OR presurgery and postsurgery. The proposed approach uses a 3D CNN for classification task with two classes: rubbing hands and other actions. The data were collected from a hospital over a span of 4 months from a single OR. Additional data were generated by simulating a situation in OR for synthetic data. The proposed approach uses I3D model, an Inception v1 architecture that inflates 2D convolutions into 3D convolutions to train three models: I3D networks for RGB, I3D networks for optical flow inputs, and a joint model. Transfer learning approach is employed by pretraining the CNN on Kinetics-400 (Carreira and Zisserman, 2017) data set. The I3D for RGB outperforms the other two models.
2.5: Integrating medical informatics and health informatics
This section surveys machine learning at the interface of medical informatics and health informatics, particularly clinical health and medical informatics (HMI).
2.5.1: Classification and regression tasks in HMI
Ralston et al. (2007) discussed patient web services in integrated delivery system portals, which provide access to EHRs as well as financing and delivery of other healthcare products such physician consultation, medical devices, and prescription medicines. These comprise point of care (POC) and post-POC services, which serve as both information retrieval and EHR compilation mechanisms that can produce data for subsequent machine learning and data science. Data integration is a key requirement of such systems (Ralston et al., 2007). As underscored by the work of Oztekin et al. (2009) on predicting heart-lung transplant survival using a combination of case data, elicited subject matter expertise, and commonsense reasoning from a basic domain theory, the most effective learning strategy is sometimes to incorporate all available data sources and then use algorithms for feature selection and construction on these. Furthermore, Holzinger and Jurisica (2014) advocated for an integrative and interactive approach to knowledge discovery and data mining in biomedical informatics (BMI)—specifically, one that is informed by objectives of both human-computer interaction and knowledge discovery in databases.
2.5.2: Reinforcement learning for HMI
Medical informatics is a field where medical practitioners have traditionally used mixed-initiative AI, especially human-in-the-loop systems. In such systems, performance elements of machine learning are used for recommendation and decision support rather than for full automation at the POC. Holzinger (2016) discussed this general practice and its rationale in modern interactive POC systems, specifically when humans in the loop are beneficial. They note that there are specific usage contexts where humans are good at spotting irrelevant features, assessing novelty and anomaly in annotation, and behavioral modeling of adversaries in security and safety contexts (such as anonymity and privacy of patient data in HMI systems). This is an important consideration for mixed-initiative HMI because the interactive machine learning (iML) framework they advocate is rooted in RL.
RL is not limited to Q-learning and its dynamic programming relatives, the temporal differences and SARSA family of algorithms, but can include complex adaptive systems such as multiarmed bandits and contextual bandits. Yom-Tov et al. (2017) explored the problem of generating physical activity reminders to diabetes patients using RL in a contextual bandit. They show a slightly positive slope in reduction of hemoglobin A1c (HbA1c) for experimental users of this system over a 6-month period, compared to a slightly negative slope for a control group.
Deep RL (DRL) systems for HMI often frame treatment tasks as optimal control problems as outlined in Section 1.4.2. Raghu et al. (2017) gave one example of such a DRL application: the administration of intravenous fluids and vasopressors in septic patients. Their DQN-based approach calculates discounted return from off-policy (historical) data rather than by directly controllable reinforcements. Liu et al. (2017) applied DQN to a high-dimension space of (about 270) actions corresponding to medicines given to acute myeloid leukemia (AML) patients who received hematopoietic cell transplantation (HCT), with the goal of preventing acute graft versus host disease (GVHD). The training data set consists of historical medical registry data for 6021 AML patients who underwent HCT between 1995 and 2007; these data are sequential but asynchronous (with standard follow-up forms collected at 100 days, 6 months, 12 months, 2 years, and 4 years). The authors demonstrated value enhancement of up to 21.4% using DQN and note that a larger training data set compiled by the Center for International Blood and Marrow Transplant Research (CIBMTR) can potentially be used for DRL.
Inverse reinforcement learning (IRL) is the problem of inferring the reward function of an observed agent, given its behavior as reflecting a policy. Yu et al. (2019c) addressed the IRL task of capturing the reward functions for mechanical ventilation and sedative dosing in ICUs from the Medical Information Mart for Intensive Care (MIMIC III), an open data set containing records of demographics, vital signs, laboratory tests, diagnoses, and medications for nearly 40,000 adult and 8000 neonatal ICU patients. The authors demonstrated that IRL from this critical care data is feasible using a Bayesian inverse Q-learning algorithm (fitted Q-iteration) applied to an MDP representation.
An MDP is also used by Yu et al. (2019a) in a direct RL application. Here, they incorporate causal factors between options of anti-HIV drugs and observed effects into a model-free policy gradient RL learning algorithm, to learn DTRs for human immunodeficiency virus (HIV). This approach is shown to facilitate direct learning of causal policy gradient parameters without requiring a model-based intermediary, a finding which has potential relevance to DRL and IRL as well.
2.5.3: Self-supervised, transfer, and active learning in HMI
Qiu and Sun (2019) applied this paradigm to RL in vision, for iterative refinement of tomographic images and in order to improve diagnostic image classification. The specific type of imaging is called optical coherence tomography.
Self-supervision is also used in other deep learning representations. Among the earliest of these to be developed were CNNs, which in HMI are designed to perform visual phenotyping. Gildenblat and Klaiman (2019) developed a Siamese network for segmentation of pathological images (e.g., into regions corresponding to stromata, tumors infiltrating lymphocytes, blood vessels, fat, healthy tissue, necrosis, etc.). Sarkar and Etemad (2020) used a CNN to learn relevant patterns from electrocardiograms for emotion recognition.
Another self-supervised learning representation is found in word embedding models in NLP. Meng et al. (2020) used BERT to classify radiology reports by urgency, learning a contextual representation rather than using more traditional sentiment analysis or other methods requiring word-level grading or subjective annotation of the training corpus.
Yet another self-supervised learning architecture is the generative adversarial network (GAN), which has been applied to create artificial images (DeepFakes) and achieve style transfer from drawings and paintings to photographic images. Tachibana et al. (2020) used a GAN to improve the electronic cleansing process to remove fecal artifacts in CT colonoscopy images.
3: Techniques for machine learning
3.1: Supervised, unsupervised, and semisupervised learning
We provide a very brief description of each of the machine learning methods that have been used in the medical informatics literature.
3.1.1: Shallow
A decision tree (Brodley and Utgoff, 1995) is a directed tree model that is used as a rule-induction system where each node represents a feature (univariate decision tree) and following a path of features (represented as nodes in the tree) leads to a specific classification.
NB (Rish et al., 2001) is a probabilistic classifier used for binary or multiclass classification problems. It is based on the poor assumption of conditional independence among the features and uses Bayes’ theorem.
ANNs (Hassoun et al., 1995) are inspired by biological neural network and the brain’s ability to process massive amount of information in parallel that allows it to recognize and classify the world around it. Researchers introduced ANN that can be created using a weighted directed graph to process information in parallel where nodes of the graph are connected together in a similar way the neurons in the brain are connected.
SVMs (Suykens and Vandewalle, 1999) use a mathematical function called kernel that transforms linearly inseparable data to linearly separable by finding the hyper-plane (decision boundary) that maximizes distance from data points on either side of the plane.
RFs (Breiman, 2001) are initially a group of decision trees where each tree randomly selects a sample from the data set. Once the trees (forest) have been built, each tree produces a class prediction, which is used in the vote for the most popular class. The majority vote is used as the final classification of an input example.
kNN (Dudani, 1976) is a supervised, nonparametric, and lazy learning algorithm that is used for classification and regression problems. A kNN implementation has three important components—training and test data set; an integer value K; and a distance metric such as Euclidean, Manhattan, or Hamming.
3.1.2: Deep
Convolutional neural networksTajbakhsh et al. (2016) explained CNNs as a special class of ANNs where each neuron from one layer does not fully connect to all neurons in the next layer. CNNs work by using convolutional layers that are mainly for detecting certain local features in all locations of their input images. A set of convolutional kernels is responsible for learning a set of local features within input images, which results in a feature mapping. Anwar et al. (2018) showed that CNNs are wildly used in medical imaging which help to achieve the following tasks: segmentation, detection and classification of abnormality, computer-aided detection or diagnosis, and medical image retrieval.
A U-Net (Ronneberger et al., 2015) is a CNN network that consists of a contracting path to capture context, and a symmetric expanding path that enables precise localization (class label is supposed to be assigned to each pixel). The architecture of the network resembles a U-shaped network of CNNs where the left side of the U-shape consists of a contracting path and the right side consists of an expansive path.
Stacked denoising autoencoders (SDAE) (Vincent et al., 2010) are multilayered denoising autoencoders that are stacked together for training purposes. These networks are trained by adding noise to the raw input, which is then fed to the first denoising autoencoder in the stack. After minimizing the loss (convergence), the information in the hidden layer (latent features) is obtained and again noise is added, and the resulting features are fed to the next denoising autoencoder in the stack. This process continues for all denoising autoencoders in the stack.
GANs (Goodfellow et al., 2014) consist of two models—a generative model and a discriminative model that are simultaneously trained. The generative model captures the data distribution by producing fake data, and the discriminative model determines if a sample comes from the real training data or from the generative model (fake data). The training goal is to maximize the probability of the discriminative model making a mistake.
RNNs (Übeyli, 2010) are a special type of neural network where the output from previous step is fed as input to the current step. The most important feature of RNN is their internal states which allow the network to have the ability of remembering information about a sequence of inputs. This is especially useful when trying any kind of prediction that relies on information that can be viewed as a sequence, such as an EEG signal.
3.2: Reinforcement learning
3.2.1: Traditional
Q-learning (Watkins and Dayan, 1992) is a simple RL algorithm that given the current state, seeks to find the best action to take in that state. It is an off-policy algorithm because it learns from actions that are random (i.e., outside the policy). The algorithm works in three basic steps: (1) the agent starts in a state and takes an action and receives a reward; (2) for the next action, the agent has two choice—either reference the Q table and select an action with the highest value or take a random action; and (3) agent updates the Q-values (i.e., Q[State, Action]). A Q-table is a reference table for the agent to select the best action based on the Q-value.
3.2.2: Deep RL
The Deep Q-Network (DQN) model by Sorokin et al. (2015) combine Q-learning (discussed earlier) with a deep CNN. The goal of this model is to train a network to approximate the value of the Q function which maps state-action pairs to their expected discounted return. The inputs to the neural network are the state variables and the outputs are the Q-values.
3.3: Self-supervised, transfer, and active learning
3.3.1: Traditional
Although the term “self-supervised” dates back to learning systems that used shallow representations rather than deep neural networks and similar representations (Hoffmann et al., 2010; Stewart et al., 2011; Roller and Stevenson, 2014), we can see from the distinct purpose and methodology used to achieve self-supervision that deep learning has emerged as a paradigm that facilitates new forms of self-supervised learning. Transfer learning has similarly been studied extensively since the advent of deep learning, but compared to self-supervision, is better understood as an independent task. A broad and comprehensive overview of the problem is provided by Torrey and Shavlik (2010).
3.3.2: Deep
AlexNet (Krizhevsky et al., 2012) is an eight-layered CNN network of five convolutional layers followed by three fully connected layers with 60 million parameters and 650,000 neurons. The authors trained this network using a subset of the popular ImageNet image corpus.
VGG (Simonyan and Zisserman, 2015) is a very deep CNN for large-scale image recognition developed as part of the Large-Scale Visual Recognition Challenge (ILSVRC).
ResNets (He et al., 2016), or residual neural networks, are a type of deep neural net architecture modeled on pyramidal cells (high-connectivity cortical neurons). To alleviate the difficulty of training deeper neural networks, ResNets contain shortcut (skip) connections that propagate activations forward by one or more layers; these activation blocks are referred to as residual blocks and this structure reformulates a baseline plain network into its counterpart residual version. ResNets have a typical depth of 50 to over 150 layers (ResNet-152), which is very big compared to VGG.
SqueezeNet (Iandola et al., 2016) is another CNN architecture which was mainly developed to achieve a similar accuracy to AlexNet, but with far less number of parameters and size. It maintains an accuracy on ImageNet data set that is comparable to that of AlexNet.
DenseNet (Huang et al., 2017) was developed based on the observation that CNNs can be deeper, more accurate, and very efficient to train if they contain shorter connections between layers close to the input and those close to the output. DenseNet consists of dense blocks where each block connects each layer to every subsequent layer in that block. The advantages of DenseNet include avoiding the vanishing-gradient problem, strengthening feature propagation, feature reuse, and reduce the number of training parameters.
4: Applications
We now survey selected popular test beds for medical predictive analytics.
4.1: Test beds for diagnosis and prognosis
The most used data set for breast cancer diagnosis is the Breast Cancer Wisconsin (Diagnostic) Data Set 2.1.1 (http://archive.ics.uci.edu/ml). This data set is publicly available from the University of California Irvine (UCI) Machine Learning Repository.
Data sets used for the task of predicting breast cancer recurrence (Abreu et al., 2016) include: (1) The Wisconsin prognostic breast cancer (WPBC) data set from the UCI ML repository (http://archive.ics.uci.edu/ml) and (2) the data set from the SEER database (SEER Incidence Database—SEER Data & Software, 2021; Choi et al., 2009).
An RNA-seq data set for cancer prediction (Xiao et al., 2018) is freely available from The Cancer Genome Atlas (TCGA) program database (The Cancer Genome Atlas Program, 2021). TCGA is a cancer genomics program that has generated over 2.5 petabytes of genomic, epigenomic, transcriptomic, and proteomic data. These data have been used to facilitate improvements in the diagnosis, treatment, and the prevention of cancer.
4.1.1: New test beds
We refer the interested reader to work by Deserno et al. (2012), Murphy et al. (2015), and Svensson-Ranallo et al. (2011), and the guidelines of the National Heart, Lung, and Blood Institute (NHLBI) for preparation of clinical study data (Guidelines for Preparing Clinical Study Data Sets for Submission to the NHLBI Data Repository, 2021).
4.2: Test beds for therapy recommendation and automation
We refer the interested reader to relevant work by Valdez et al. (2016), Son and Thong (2015), and Thong et al. (2015).
4.2.1: Prescriptions
We refer the interested reader to work by Galeano and Paccanaro (2018), Kushwaha et al. (2014), Zhang et al. (2015), Bao and Jiang (2016), Bhat and Aishwarya (2013), and Guo et al. (2016).
4.2.2: Surgery
We refer the interested reader to relevant work by Ciecierski et al. (2012), Petscharnig and Schöffmann (2017), Wang and Fey (2018), and Shvets et al. (2018).
5: Experimental results
5.1: Test bed
As an experimental example, we include a test bed introduced by Fanconi (2019) in this chapter. This consists of a subset of 3600 pictures extracted from the data set in the International Skin Imaging Collaboration Archive (ISIC), which contains more than 23,000 mole pictures. The test bed contains balanced cases of malignant (1) and benign (0) skin moles. Here, we present a simple performance comparison between different pretrained DLAs that classify a skin mole as being malignant or benign.
This data set has been used by several recent publications (Zhang et al., 2019; Tschandl et al., 2019; Nida et al., 2019; Mahbod et al., 2019).
5.2: Results and discussion
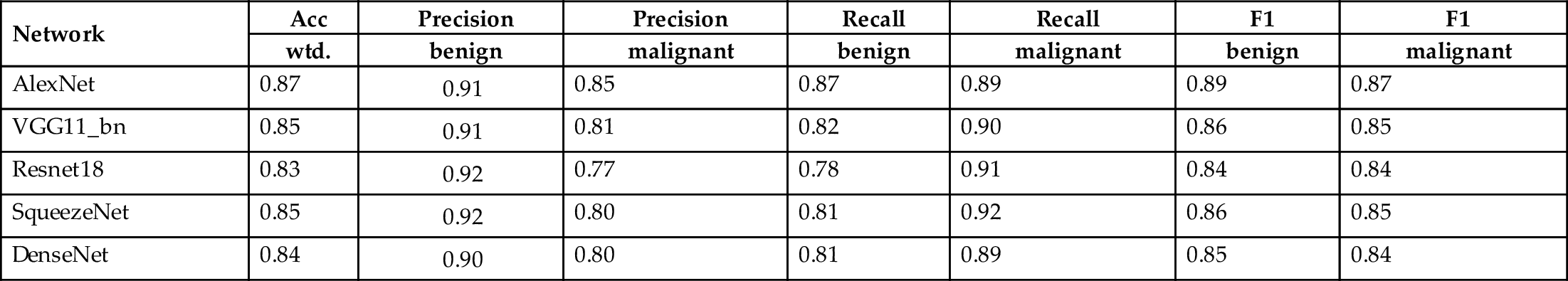
We use weighted accuracy, precision, recall, and F1-score to test the performance of the DLAs. Table 1 shows the performance of the different DLAs used in this comparison. In terms of weighted accuracy, AlexNet have achieved the best accuracy. SqueezeNet and Resnet-18 achieved the best precision score when classifying a mole as benign, but the worst precision score when classifying a mole as malignant. AlexNet has the best precision score when classifying a mole as malignant and the best F1-score as well. We can also see that the difference in the performance of the DLAs is only marginal and we believe that different data set would yield different performance between the tested DLAs.
Table 1
| Network | Acc | Precision | Precision | Recall | Recall | F1 | F1 |
|---|---|---|---|---|---|---|---|
| wtd. | benign | malignant | benign | malignant | benign | malignant | |
| AlexNet | 0.87 | 0.91 | 0.85 | 0.87 | 0.89 | 0.89 | 0.87 |
| VGG11_bn | 0.85 | 0.91 | 0.81 | 0.82 | 0.90 | 0.86 | 0.85 |
| Resnet18 | 0.83 | 0.92 | 0.77 | 0.78 | 0.91 | 0.84 | 0.84 |
| SqueezeNet | 0.85 | 0.92 | 0.80 | 0.81 | 0.92 | 0.86 | 0.85 |
| DenseNet | 0.84 | 0.90 | 0.80 | 0.81 | 0.89 | 0.85 | 0.84 |
6: Conclusion: Machine learning for computational medicine
This chapter has introduced machine learning tasks for diagnostic medicine and automation of treatment, comprising supervised, self-supervised, and RL. After surveying task definitions, we outlined existing methods for diagnosis, predictive analytics, therapy recommendation, automation of treatment, and integrative HMI. We now look forward to current and continuing work in applied machine learning for medical applications.
6.1: Frontiers: Preclinical, translational, and clinical
Machine learning has developed into a cross-cutting technology across areas of medical informatics, from theory, education, and training (preclinical) aspects to the observation and treatment of patients (clinical computational medicine), to translational medicine, which aims toward bridging experimental tools and treatments to deployed ones used in clinical practice. For preclinical surveys, we refer the interested reader to the following: Prashanth et al. (2016), who discussed experimental Parkinson’s treatments (Prashanth et al., 2016; Kolachalama and Garg, 2018) surveyed machine learning as a subarea of AI, particularly in medical education (Kolachalama and Garg, 2018); and Bannach-Brown et al. (2018) reviewed natural language learning and text mining in medical informatics. Ravì et al. (2016) surveyed deep learning in translational health informatics; Weintraub et al. (2018) reviewed translational text analytics; and Shah et al. (2019) broadly examined machine learning and AI in translational medicine, from sensors and medical devices to drug discovery. Finally, Savage (2012) provided an early survey of big data for improving clinical medicine, while Char et al. (2018) addressed ethical considerations.
6.2: Toward the future: Learning and medical automation
As of this writing in 2020, the coronavirus disease SARS-Cov-2 is of pervasive interest as a highly time-critical use case of intelligent systems for improved diagnosis and epidemiological modeling, as discussed by Randhawa et al. (2020), and for understanding etiology toward vaccine discovery, as discussed by Alimadadi et al. (2020). The (Stanford HAI (2021)) conference surveys broad methods for combatting Covid-19 and future pandemics. More generally, the fields of diagnostic medicine and predictive analytics have grown significantly since the early work of Kononenko (2001) and the health informatics applications of Pakhomov et al. (2006), but there are deep technical gaps and significant challenges remaining before the “eDoctor” envisioned by Handelman et al. (2018) becomes a reality. Meanwhile, AI safety concerns, as mapped out by Yampolskiy (2016) in both historical and anticipatory contexts of risks and remedies, loom large.
