4.3 Results
4.3.1 Community Maps
The communities retrieved for each subnetwork are listed in Table 4.1, and Figure 4.2 describes the comparative algorithmic performance through the maximal values achieved by Q for each PIN. Then, Figures 4.3–4.6 represent instead examples of community maps computed for the relevant subnetworks, that is, cPIN, cePIN, cePIN-1, and cePIN-2, respectively. A characterization for the module maps is obtained by the cell cycle phases, that is, the S (synthesis, DNA replication) and M (mitosis, chromosome separation) phases together with the remaining G1 and G2 interphases. For both cePIN-1 and cePIN the corresponding KEGG pathways have also been reported as supplementary data (SD).
Table 4.1 Detected Communities (Relatively Small Ones)

Table 4.2 Number of MCODE-Detected k-Cores at k Ranging Between Minimum of 2 and Maximum of 13 Across PIN.
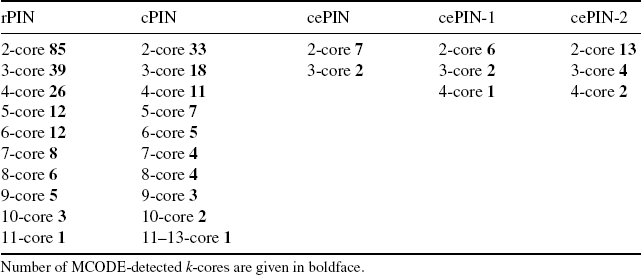
Figure 4.3 Community maps: (a) cPIN and (b) cePIN-1. The thick arrows point out the indicated hubs.
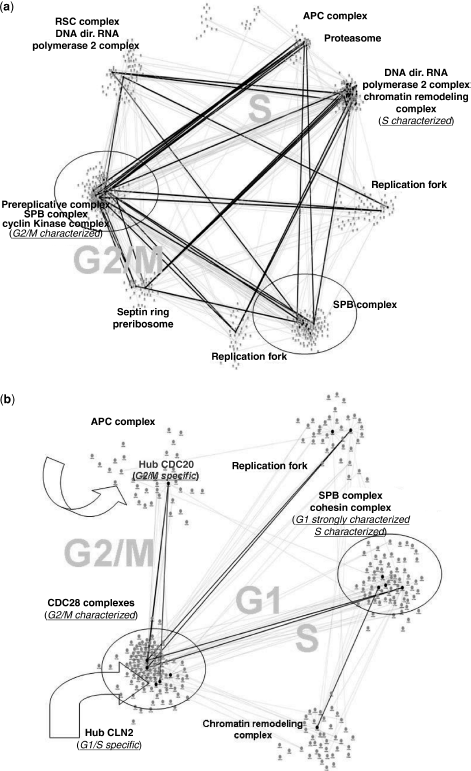
Figure 4.4 Community maps: (a) cePIN and (b) cePIN-2.
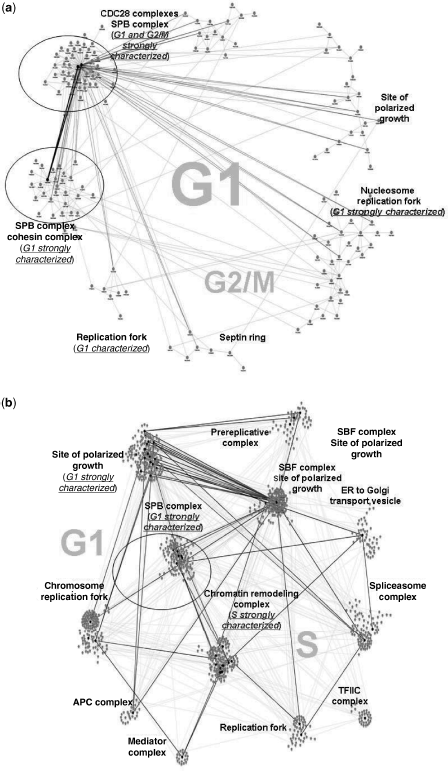
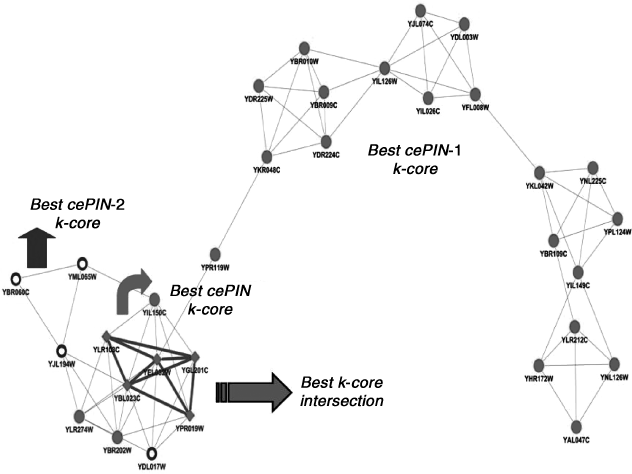
Figure 4.5 Best k-core cPIN.
Figure 4.6 Best k-core intersection from cePIN, cePIN-1, and cePIN-2.
4.3.1.1 Analysis
The analysis of the maps has been pursued according to some simple rules aimed to provide useful interpretation. As a first rule, compact maps have been formed by postprocessing the retrieved communities. In particular, hub proteins have been emphasized due to their relevant role in terms of network stability. Concerning their definition, since a connectivity threshold for distinguishing a protein hub is required but is not unique, after some tests we have retained useful hubs with at least 15 connected proteins for our purposes.
Thus, the resulting community maps are hub-centered, and allow for a coarse-grained evaluation. We compared various hubs providing community characterization with a balance between module cohesiveness and cross-links then validated through annotation. The visualized communities show darker nodes representing the hub proteins, and thick links emphasizing “hub-hub” cross-connections, together with lighter links connecting hubs to nonhub proteins. All the other links have been hidden (examples with complete associations are reported in SD).
The comparative examination of the community maps suggests some remarks. In Figure 4.3a the map obtained from cPIN with just one driver (interactions) delivers a quite redundant map, and extra cell cycle related communities appear due to possible algorithmic errors. In the other maps, it is crucial to check the contribution from coexpression-induced dynamics. In particular, Figure 4.3b reports the cePIN-1 map; only half of the previously shown communities in cPIN are left.
The SPB or spindle pole body complex is the microtubule organizing center in the yeast cells, while the Cohesin complex is responsible for binding the sister chromatids during synthesis through the G2 phase and into M phase. The CDC28 complexes appear too, with the cdk or cyclin-dependent kinase that phosphorylates a variety of target substrates, together with the APC or anaphase-promoting complex (cyclosome) that promotes metaphase–anaphase transition by ubiquitinating its specific substrates (such as mitotic cyclin and anaphase inhibitor), then subsequently degraded by the proteasome.
One of the communities is characterized by both the SPB and the Cohesin, while another linked community is characterized by the CDC28 complexes. Also, communication between the latter complexes and the APC-characterized community occurs in hub–hub style between cdc28 and clb2 with cdc20, and also in non hub–hub style through the hub cln2 protein. In particular, clb2 is a B-type cyclin that activates cdc28 to promote the transition from G2 to M phases, and then its degradation is activated by the APC/Cdc20 to promote mitotic exit [25].
Then, the hub cdc28 links to SPB [26] and with reference to the same community pair, the hub cdc5 that is involved in regulation of sister chromatid separation [27] interacts with the nonhub scc1 cohesin protein. Finally, the hub sth1 protein, which is a component of the RSC or chromatin remodeling complex associating the cohesin with centromeres and chromosome arms [28], also interacts with the nonhub scc1 cohesin protein.
The KEGG pathway map reported in the SD (KEGGSDf1.jpg file) localizes (marked by circles) especially the APC closeness to cdc20, and the Cohesin connection to CDC28. In terms of phases, there appears a mix of them involving G1, G2/M, and S. In particular, cdc20 in G2/M phase justifies the hub–hub cross-links between APC and CDC28. Then, the G1 and S specificity of both the SPB and Cohesin complexes comes also from cln2, which is linked to APC too.
By looking at Figure 4.4a, the first remark is for the G1 phase that strongly characterizes the cePIN community map. A community is characterized by both CDC28 and SPB, with the latter also sharing a community with the Cohesin complex. The two main modifications that can be observed refer to the hub cdc20 that becomes a nonhub protein, and APC (activated by cdc20) that disappears (i.e., due to inactivated cdk in S/M phases that allows the cell to exit from mitosis). The strong G1-characterization can justify the absence of the link between APC and cdc20 as the latter acts in M and not G1 phase where it acts with another protein. As both Cohesin and SPB remain, the presence of the latter in G1 is possibly justified by the SPB duplication during this phase (see Refs. 29,30).
Notably, the cePIN modularity map appears reduced, and highly G1-specific. As the hub proteins have been found in just two communities, a very localized map results from the highest possible influence of peak coexpression signatures. In SD (KEGGSDf2.jpg and KEGGSDf3.jpg files), the associated KEGG pathways are reported for the two main communities.
The examination of cePIN-2 in Figure 4.4b reveals a map significantly denser than before. Quite clearly, the dimensions of each PIN are different, but it is important to underline the following aspects. This final transition naturally allows for introducing extra communities that are not related to the cell cycle, for instance, mediator, spliceasome, ER to Golgi transport vesicle, TFIIC complexes. Thus, a redundancy of communities appears compared to cePIN-1, as it was expected from the introduction of non cell cycle proteins. In terms of phases, G1 and S are both present, but with lesser characterization than before.
Overall, the community maps reflect quite well the co-expression dynamics, and in particular identify for cePIN a phase-specific modularization due to peak signatures. When relaxing the constraint of peak–peak protein interaction, the distinct impact of biological process on modularization emerges, which appears from cePIN-1. Then, when all proteins are considered as in cePIN-2, the community map substantially diversifies from strictly cell cycle based modules. Modularity is thus shaped by the drivers and the dynamics characterized accordingly.
4.3.2 Core Structures
The implementation of MCODE requires that some parameters are set in order to compute the network-scoring values. For instance, we set degree cutoff = 2 and node score cutoff = 0.2, and then proceeded by fixing other values, such as haircut = true, fluff = false; k-Core = 2; max. depth from seed = 100. The list of the retrieved k-cores has been reported in Table 4.2. We have then comparatively evaluated the results based on these k-cores, and reported them in Figures 4.4–4.6.
We looked at k-core modularity according to two different strategies. First, we have considered the most important structure provided by the method, and thus the best k-cores for each PIN. In particular, the “best k-core intersection” has been identified and targeted. The best k-core of cPIN has been reported apart in Figure 4.4 as it differentiates from the other cases in terms of data sources. Since the best k-cores (Fig. 4.5) represent modules with the highest scores, they can be considered locally optimal at the network scale. When annotation is done, the best cePIN k-core indicates a little module with highly coexpressed genes, while the proteins address the MCM complex involved in initiation and regulation of DNA replication. Also the best cePIN-2 k-core includes the latter complex, but through a larger module annotating for the prereplicative complex. Finally, the best cePIN-1 k-core results the most structured and extended one.
We observe the presence of an area of intersection that involves the whole cePIN core, and appears totally included by the other two structures. The most structured k-core with regard to cell cycle belongs to cePIN-1, and is also determined by co-expression. When we relax the requirement that all proteins refer to the cell cycle, as with cePIN-2, then the best k-core structure reduces drastically and converges to that observed for cePIN by corresponding to the bottom-left region of the cePIN-1 k-core (except for a few newly established associations).
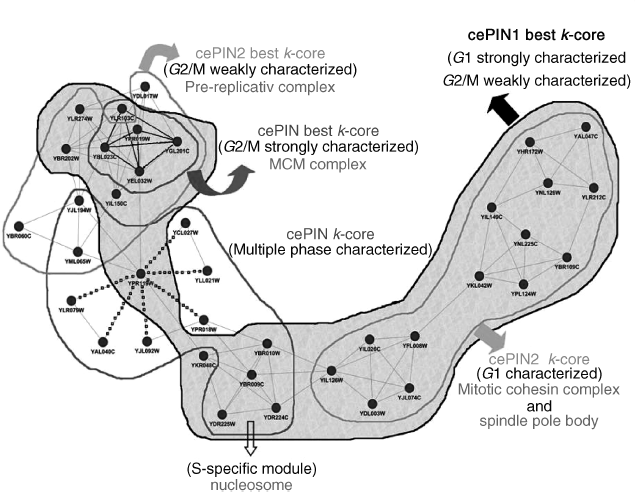
Second, considering as in Figure 4.6 the whole-layered coreness structure requires that for each PIN the innermost or most densely connected core is computed, which equivalently requires the exploration of the sequence of cores at all k, and including therefore, the best k-core. In some cases, as for cePIN-1, we observed a convergence between the best and the innermost k-cores, although the same convergence was not observed in the other two cases. The annotation is reported for the marked regions defining cores, and an interesting aspect regards the presence of a module with dotted links between nodes that reports checkpoints of the cell cycle through the cyclins.
The analysis of Figure 4.7 involves envelopes to visualize different cores belonging to the various PIN, and distinguish between the best and the innermost k-cores at varying k. Then, all characterizing phases have been indicated for each k-core. It results that the cePIN is strongly G2/M characterized (particularly through the best k-core) and presents an S-specific module, while cePIN-1 is strongly G1-characterized (and marginally G2/M-characterized too), and cePIN-2 is similar to cePIN-1 but less characterized.
Figure 4.7 Best k-cores and innermost k-cores.
We stress the fact that both strategies consider exclusively the inherent coreness structure detected by MCODE, but exclude other possible structures embedded at different resolutions [31]. In particular, we have emphasized how the cohesiveness of cores depends by the module drivers, and have evaluated the extent by which they contribute to form and differentiate cores and communities in each PIN. Overall, the clique-based modular organization is influenced by both drivers. The effect of accounting for their simultaneous presence, as in cePIN, instead of considering them more separately, is that more compact protein cores are found as expected.
4.3.3 On Network Entropy
The concept of network entropy is interesting for several reasons (see the review and developments presented extensively in Ref. [32] and also including characterizations in biology). Entropy as a measure of uncertainty can characterize PIN at both global and local levels. In particular, when local dynamics are investigated through modules, entropy can be used to corroborate the reliability of connectivity maps that are derived from modules. This strategy is pursued in our work in relation with the modular structure already extracted, that is, cores and communities.
Estimating entropies from finite samples remains in general a complicated task due to statistical fluctuations, and this limitation holds also for sampled networks. In the limit of sample size, when steady-state conditions are achieved, the Shannon entropy associated with the network distribution p(k) is given by
(4.3) ![]()
The same formula has been applied in this work to approximately investigate the uncertainty levels referred to cores and communities computed over the cell cycle related subinteractomes. As a result, Figures 4.8–4.10 have been generated. As a first observation, communities are larger modules compared to cores, and thus show in general bigger values of the estimated entropy. Protein hubs are largely responsible for the latter values, as it appears from the top plots in the first two figures (the last one has a long list of hubs that we report apart).
Figure 4.8 cePIN entropies with hubs emphasized for both (a) cores and communities and (b) sorted by size.
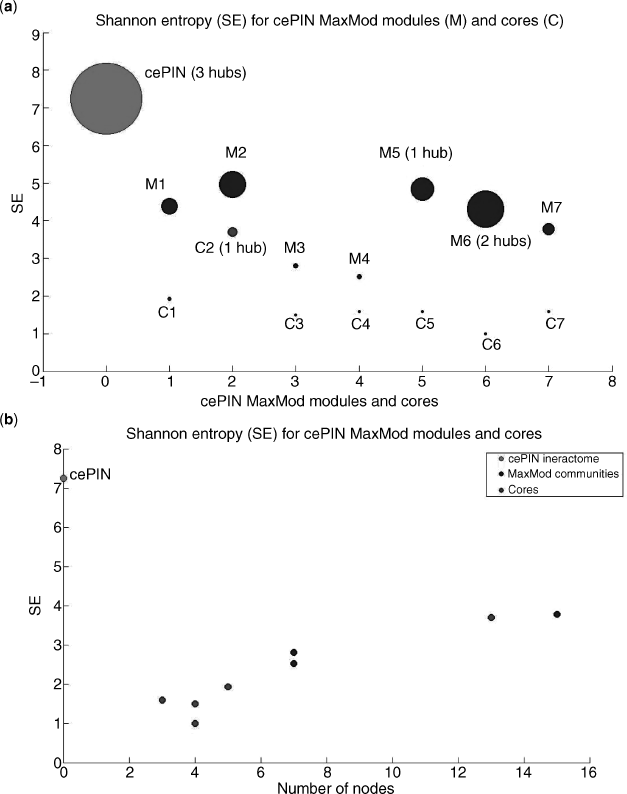
Figure 4.9 cePIN1 entropies with hubs emphasized for (a) both cores and communities and (b) sorted by size.
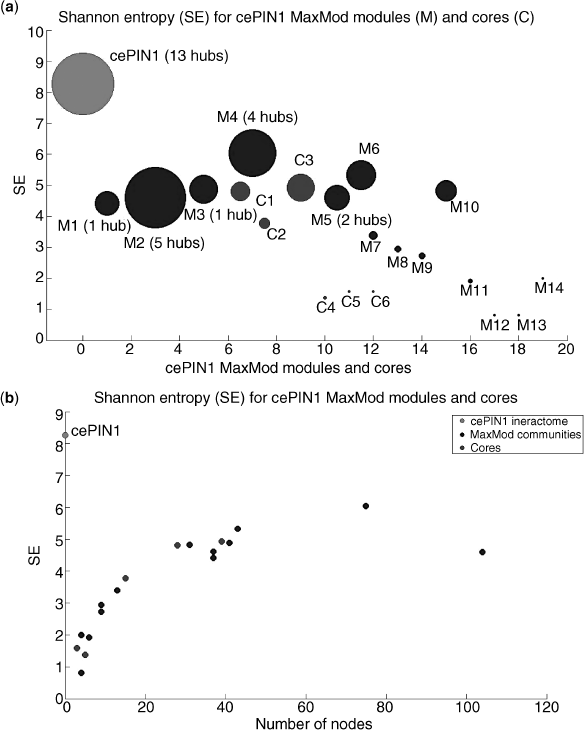
Figure 4.10 cePIN2 entropies with hubs emphasized for both (a) cores and communities and (b) sorted by size.
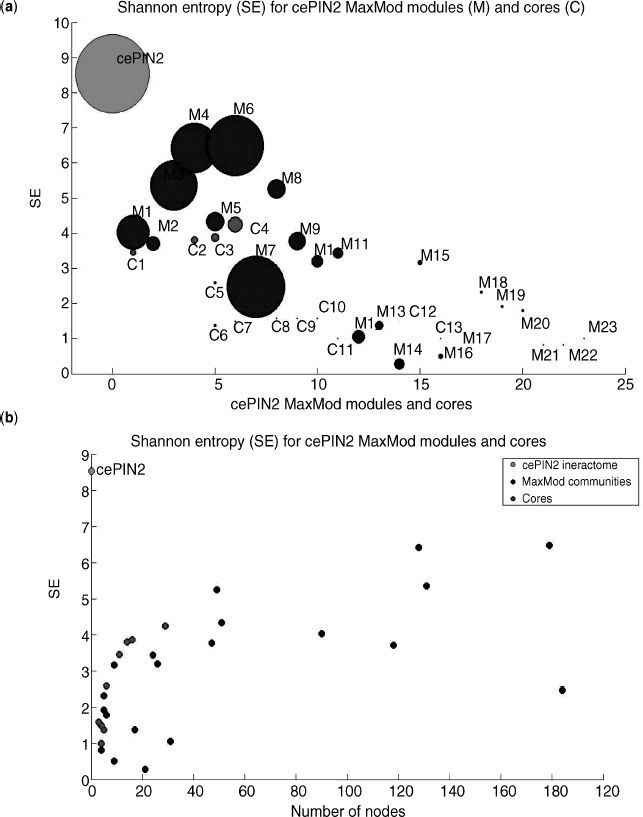
Notably, when the number of nodes is considered in the X axis (see bottom plots), we observe that cores shift toward the origin (i.e., relatively smaller cores for given entropy levels) the more the cell cycle effects are relaxed. This means that the cell cycle dynamics may be monitored by measuring entropy in relation to the module sizes, and what we may observe reflects the fact that random effects play a major role under relaxed conditions.
