Putting a model into production is one of the most challenging tasks in the data science world. It is one of those last-mile problems that persists in many organizations. Although there are many tools for managing workflows, as the organization matures its needs change, and managing existing models can become a herculean task. When we take a step back and analyze why it is so challenging, we can see that it is because of the structure that exists in most organizations. There is an engineering team that maintains the production platform. There is a gap between the data science toolset and the production platforms. Some of the data science work can be developed in Jupyter Notebook, with little consideration given to the cloud environment. Some of the data flows are created locally with limited scaling. Such applications tend to falter with large amounts of data. Best practices that exist in the software development cycle don’t stick well with the machine learning lifecycle because of the variety of tasks involved. The standard is mainly defined by the data science team in the organization. Also, rapid developments in the field are leaving vacuums with respect to the management and deployment of models.
The goal of this chapter is to explore the tools available for managing and deploying data science pipelines. We will walk you through some of the tools available in the industry and demonstrate with an example how to construct an automated pipeline. We will use all the concepts that we have learned so far to construct these pipelines. By the end of this chapter, you will have learned how to build the machine learning blocks and automate them in coherence with custom data preprocessing.
MLflow. We will introduce MLflow as well as its components and advantages, followed by a demonstration. In the first half, our focus will be on the installation and introduction of concepts. In the second half, we will focus on implementing a model in MLflow.
Automated machine learning pipelines. We will introduce the concept of designing and implementing automated ML frameworks in PySpark. The first half of the section is heavily focused on implementation, and the second half focuses on the outputs generated from the pipeline.
MLflow
With the knowledge obtained from previous chapters, we know how to build individual supervised and unsupervised models. Often times we would try multiple iterations before picking the best model. How do we keep track of all the experiments and the hyperparameters we used? How can we reproduce a particular result after multiple experiments? How do we move a model into production when there are multiple deployment tools and environments? Is there a way to better manage the machine learning workflow and share work with the wider community? These are some of the questions that you will have after you are comfortable building models in PySpark. Databricks has built a tool named MLflow that can gracefully handle some of the preceding questions while managing the machine learning workflow. MLflow is designed to be open interface and open source. People often tend to go back and forth between experimentation and adding new features to the model. MLflow handles such experimentation with ease. It is built around REST APIs that can be consumed easily by multiple tools. This framework also makes it easy to add existing machine learning blocks of code. Its open source nature makes it makes it easy to share the workflows and models across different teams. In short, MLflow offers a framework to manage the machine learning lifecycle, including experimentation, reproducibility, deployment, and central model registry.

MLflow components
Tracking: Aids in recording, logging code, configurations, data, and results. It also gives a mechanism to query experiments.
Projects: This component of MLflow helps in bundling the machine learning code in a format for easy replication on multiple platforms.
Models: This assists in deploying machine learning code in diverse environments.
Model Registry: This component acts like a repository in which to store, manage, and search models.
This framework opens the tunnel for data engineers and data scientists to collaborate and efficiently build data pipelines. It also provides the ability to consume and write data from and to many different systems. It opens up a common framework for working with both structured and unstructured data in data lakes as well as on batch streaming platforms.
This may look abstract for first-time users. We will walk you through an example of how these components can be useful and implemented. For this purpose of illustration, let us take the same bank dataset we used in Chapter 6.
MLflow Code Setup and Installation
Code/script changes
Docker side changes and MLflow server installation
Code changes are minimal. We have to add a few MLflow functions to existing machine learning code to accommodate MLflow.
Regular code works well for building a simple random forest model, but what if I want to know the Receiver Operating Characteristic (ROC)/other metrics (accuracy, misclassification, etc) by changing the input variables or any hyperparameter settings? Well, if it’s a single change we can record each run in Excel, but this is not an efficient way of tracking our experiments. As data science professionals, we run hundreds of experiments, especially in the model fine-tuning phase. Is there a better way of capturing and annotating these results and logs? Yes, we can do it via the MLflow tracking feature. Now, let’s rewrite a simple random forest model code, adding MLflow components. Changes in the code are highlighted in bold.
We will save the following code as .py and run via Docker after installing MLflow.
In the preceding snippet of code, we added log_param and log_metric to capture the pieces of information we want to keep tabs on. Also note we are logging the model using the mlflow.spark.log_model function , which helps in saving the model on the MLflow backend. This is an optional statement, but it is handy if you want to register the model from MLflow. With minimal changes in code, we are able to accommodate an existing model with MLflow components. Why is it so important? Tracking the change in parameters and metrics can become challenging with hundreds of experiments staged over multiple days. We will need to save the preceding code as a Python file. We will be using a Spark submit to execute this PySpark code. In this illustration, we saved it as Chapter9_mlflow_example.py.
MLflow by default uses port 5000.
HDFS
NFS
FTP server
SFTP server
Google Cloud storage (GCS)
Azure Blob storage
Amazon S3
Storage Types
Storage Type | URI Format |
|---|---|
FTP | ftp://user:pass@host/path/to/directory |
SFTP | sftp://user@host/path/to/directory |
NFS | /mnt/nfs |
HDFS | hdfs://<host>:<port>/<path> |
Azure | wasbs://<container>@<storage-account>.blob.core.windows.net/<path> |
Amazon S3 | s3://<bucket>/<path> |
There are additional settings needing to be authenticated for a few storage types. For S3, credentials can be obtained from environment variables AWS_ACCESS_KEY_ID and AWS_SECRET_ACCESS_KEY or from the IAM profile.
MLflow User Interface Demonstration

MLflow UI
As you can see, the user interface has all the information formatted into experiments and models. We can make any annotations or notes using the notes options pertaining to each of the runs within each experiment. This framework gives us the ability to manage and run multiple experiments with multiple runs. We also have an option to filter the runs based on parameter settings. This is extremely useful in a machine learning lifecycle because, unlike the software development lifecycle, we tend to iterate between old and newer versions based on the stability and accuracy of the models.
There is also detailed row-level information on each run. We can drill down and compare this information across many experiments. For our illustration, we have only used a single metric, but in a real-world scenario, we may also want to capture accuracy, misclassification, lift, and KS statistics for each run. We could then compare and sort each iteration based on a metric that is acceptable based on business need.

MLflow UI comparison of runs

MLflow UI comparison window

MLflow UI Run window
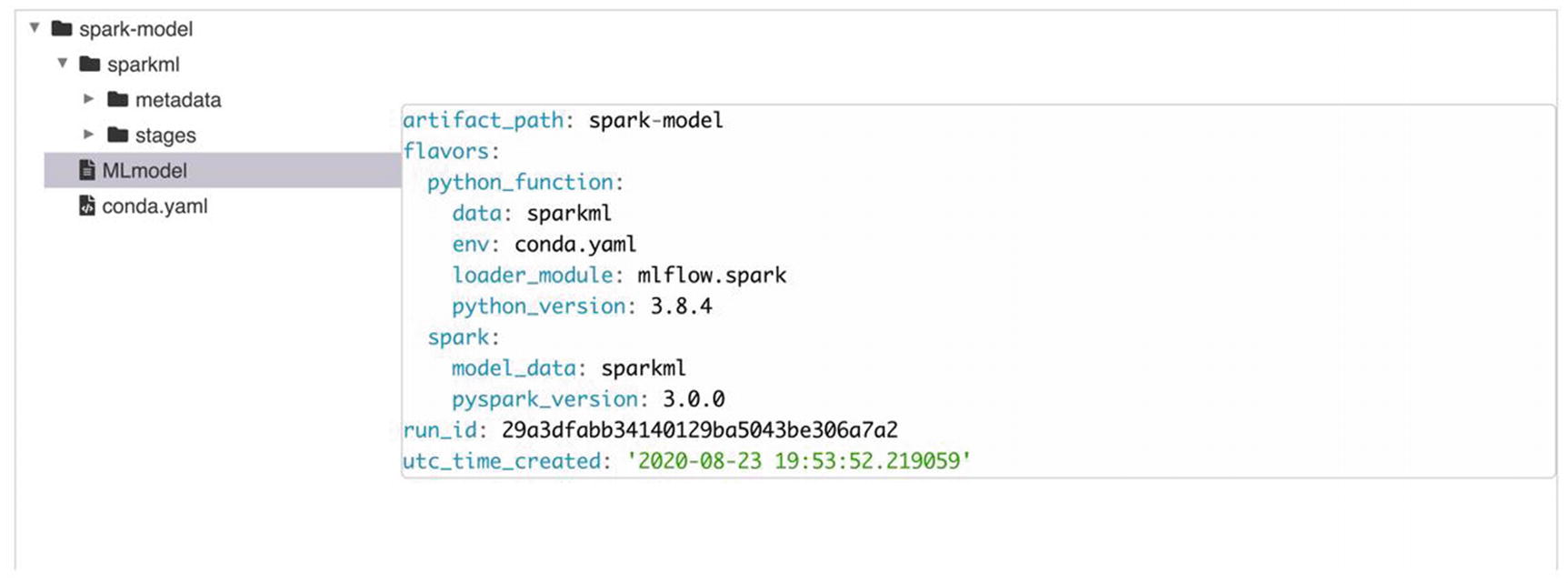
MLflow UI model information
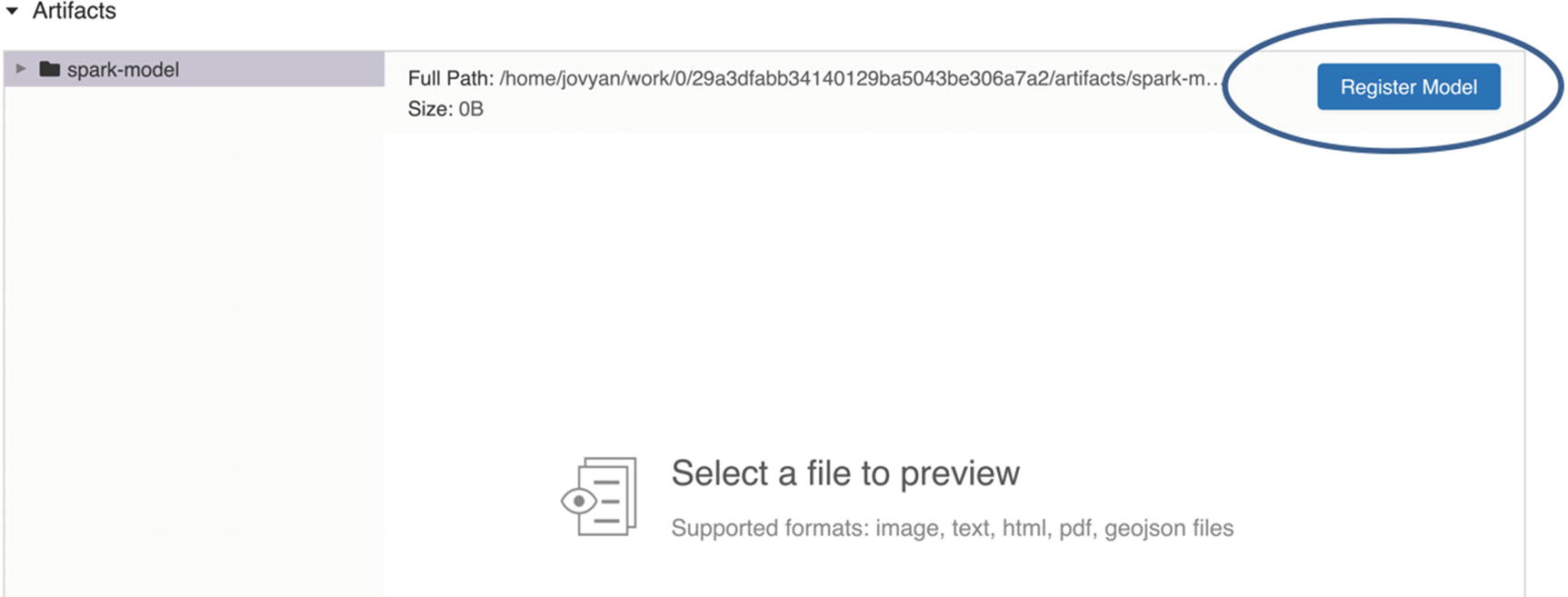
MLflow UI model registration
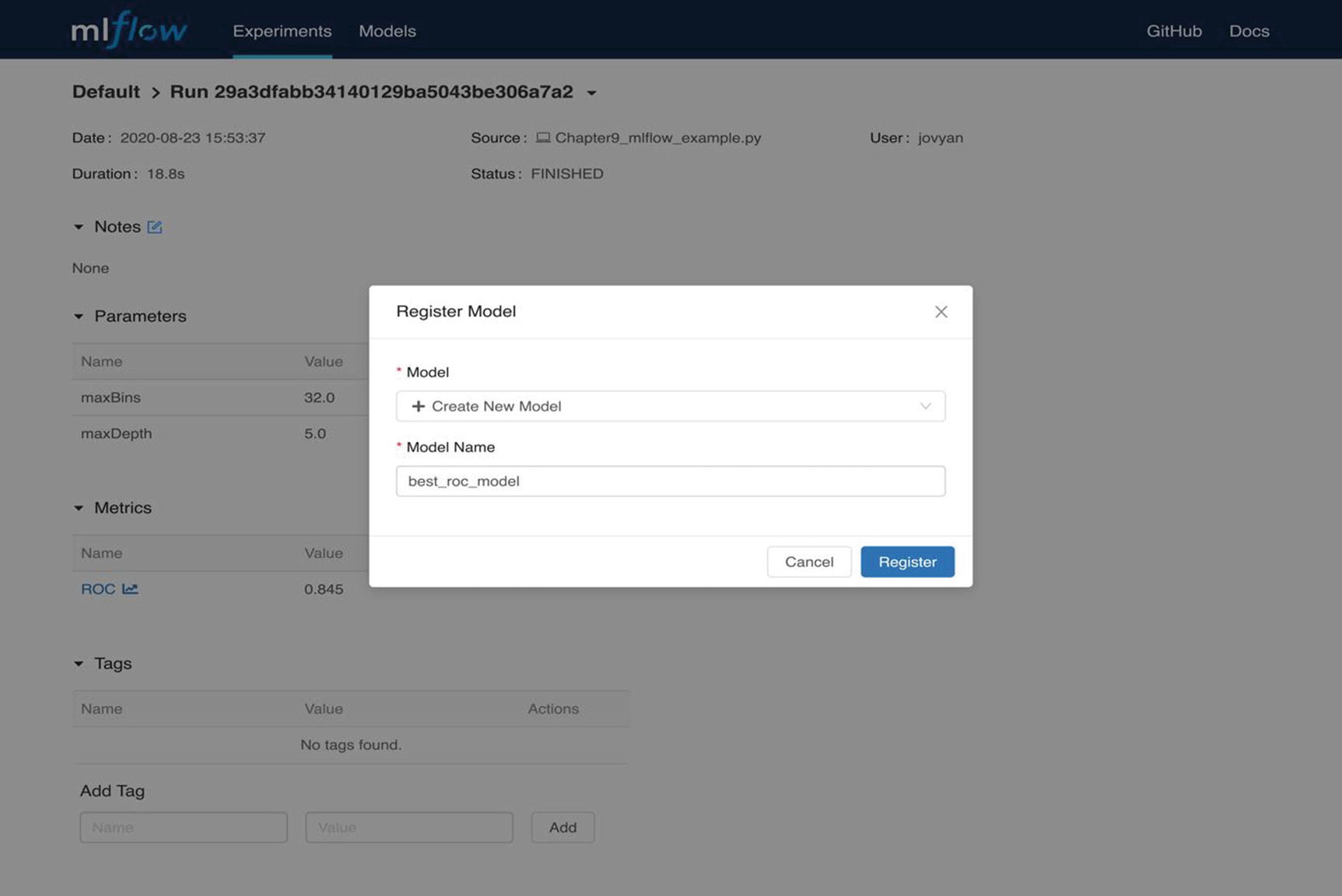
MLflow UI Registration window

MLflow Models tab
Staging
Production
Archived

MLflow model environments
This helps in effectively managing the model lifecycle by moving models to different environments as we transition from model development to the end of the model lifecycle.
All the preceding content and codes are specified in Spark. There are multiple other flavors that MLflow supports. We can serialize the preceding pipeline in the mleap flavor, which is a project to host Spark pipelines without a Spark context for smaller datasets where we don’t need any distributed computing. MLflow also has the capability to publish the code to GitHub in the MLflow project format, making it easy for anyone to run the code.
Automated Machine Learning Pipelines
The machine learning lifecycle is an iterative process. We often tend to go back and forth tuning parameters, inputs and data pipelines. This can quickly get cumbersome with the number of data management pipelines. Creating automated pipelines can save a significant amount of time. From all the earlier chapters we have learned data manipulations, algorithms, and modeling techniques. Now, let us put them all together to create an automated PySpark flow that can generate baseline models for quick experimentation.
Metadata
Column | Description |
|---|---|
RowNumber | Identifier |
CustomerId | Unique ID for customer |
Surname | Customer's last name |
Credit score | Credit score of the customer |
Geography | The country to which the customer belongs |
Gender | Male or female |
Age | Age of customer |
Tenure | Number of years for which the customer has been with the bank |
Balance | Bank balance of the customer |
NumOfProducts | Number of bank products the customer is utilizing |
HasCrCard | Binary flag for whether the customer holds a credit card with the bank or not |
IsActiveMember | Binary flag for whether the customer is an active member with the bank or not |
EstimatedSalary | Estimated salary of the customer in dollars |
Exited | Binary flag: 1 if the customer closed account with bank and 0 if the customer is retained |
Pipeline Requirements and Framework
KS: Kolmogorov-Smirnov test is a measure of separation between goods and the bads. Higher is better..
ROC: (ROC) Curve is a way to compare diagnostic tests. It is a plot of the true positive rate against the false positive rate. Higher is better.
Accuracy: (true positives+true negatives)/ (true positives+true negatives+false positives+false negatives). Higher is better.
Module should be able to handle the missing values and categorical values by itself.
We also prefer the module to take care of variable selection.
We also want this module to test out multiple algorithms before selecting the final variables.
Let the module compare different algorithms based on preferred metric and select the champion and challenger models for us.
It would be handy if it could remove certain variables by prefix or suffix or by variable names. This can power multiple iterations and tweak input variables.
Module should be able to collect and store all the output metrics and collate them to generate documents for later reference.
Finally , it would be nice if the module could save the model objects and autogenerate the scoring codes so we can just deploy the selected model.

CRISP–DM
Data manipulations
Feature selection
Model building
Metrics calculation
Validation and plot generation
Model selection
Score code creation
Collating results
Framework to handle all the preceding steps
Data Manipulations
Missing value percentage calculations
Metadata categorization of input data
Handling categorical data using label encoders
Imputing missing values
Renaming categorical columns
Combining features and labels
Data splitting to training, testing, and validation
Assembling vectors
Scaling input variables
Feature Selection
Model Building
Metrics Calculation
Validation and Plot Generation
Model Selection
Score Code Creation
Collating Results
Framework
This framework file combines all eight preceding modules to orchestrate them in a logical flow to create the desired output. It treats the data and performs variable selection. It builds machine learning algorithms and validates the models on holdout datasets. It picks the best algorithm based on user-selected statistics. It also produces the scoring code for production.
Files and their functions in the Automation Framework
Module | Filename |
|---|---|
Data manipulations | data_manipulations.py |
Feature selection | feature_selection.py |
Model building | model_builder.py |
Metrics calculation | metrics_calculator.py |
Validation and plot generation | validation_and_plots.py |
Model selection | model_selection.py |
Score code creation | scorecode_creator.py |
Collating results | zipper_function.py |
Framework | build_and_execute_pipe.py |
After the execution is completed, all the data will be stored in /home/jovyan/.
Nice job! We have successfully built an end-to-end automation engine that can save you a significant amount of time. Let’s take a look at the output this engine has generated.
Pipeline Outputs
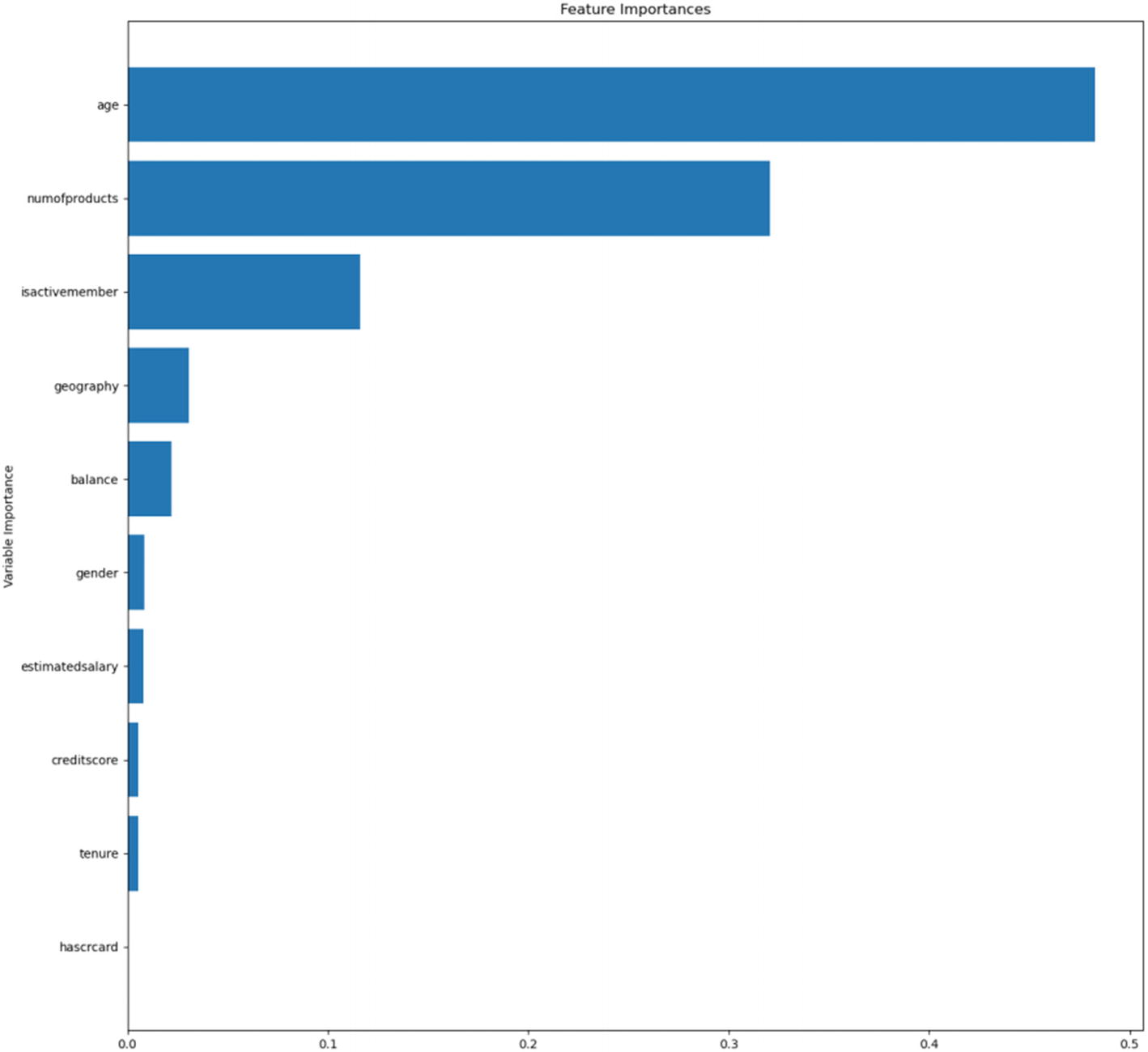
Automated model feature importances
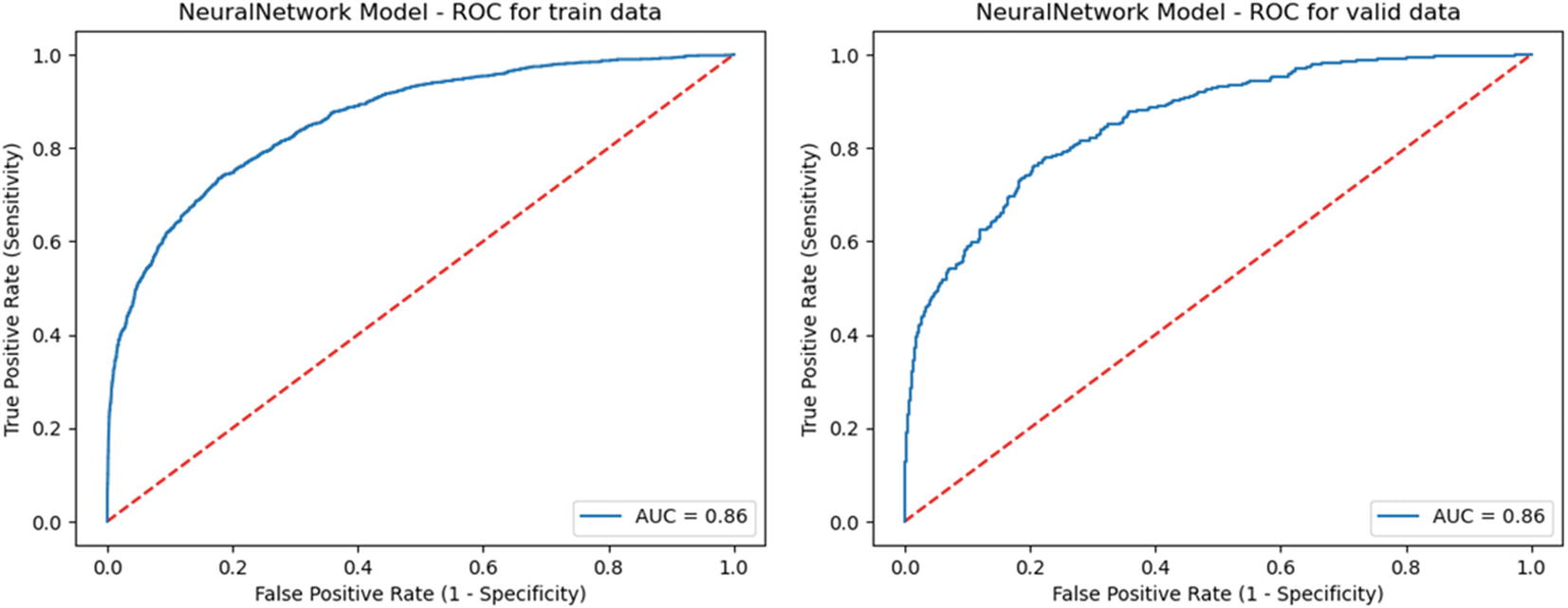
Automated model ROCs
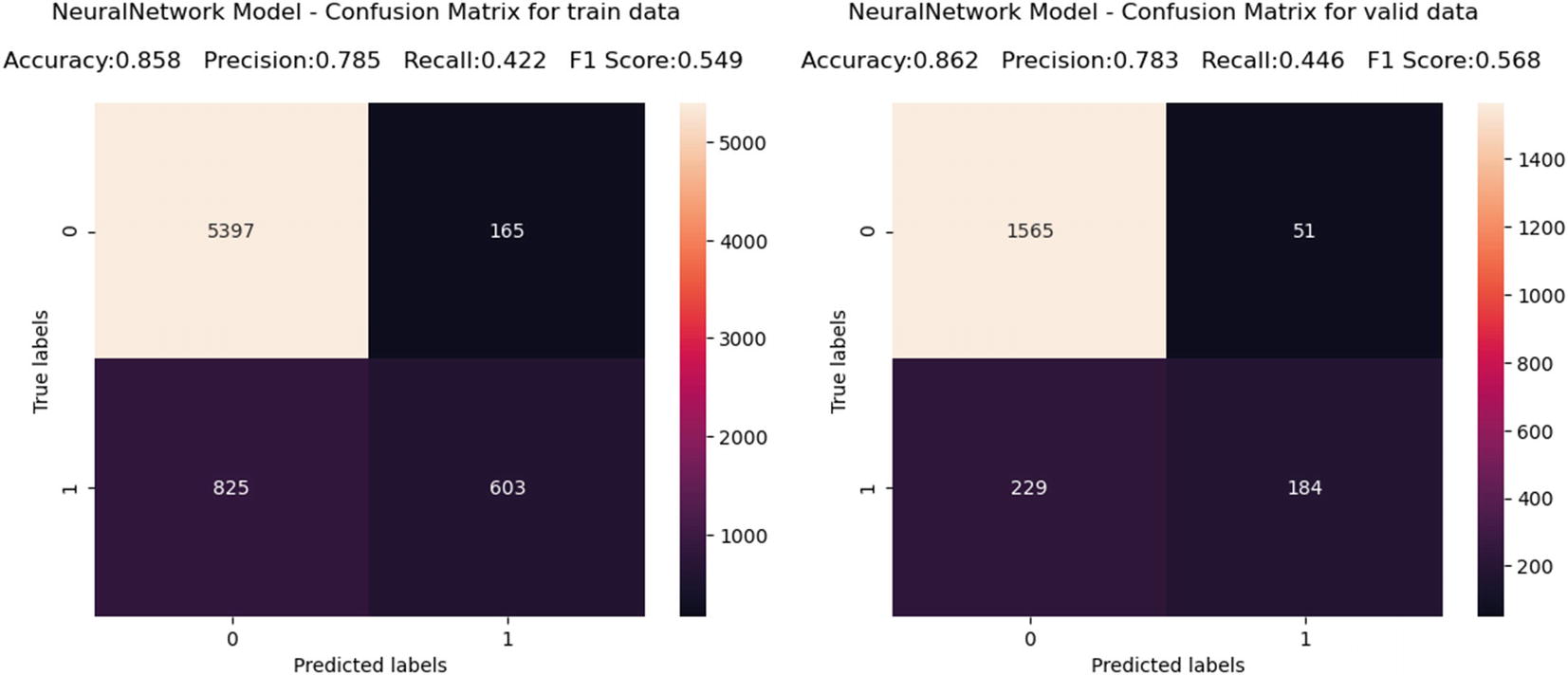
Automated model — Confusion matrix
Combined Metrics
model_type | roc_train | accuracy_train | ks_train | roc_valid | accuracy_valid | ks_valid |
|---|---|---|---|---|---|---|
neuralNetwork | 0.86171092 | 0.858369099 | 55.5 | 0.857589 | 0.862000986 | 54.43 |
randomForest | 0.838162018 | 0.860228898 | 50.57 | 0.835679 | 0.856086742 | 50.48 |
logistic | 0.755744261 | 0.81230329 | 37.19 | 0.754326 | 0.807294234 | 37.31 |
gradientBoosting | 0.881999528 | 0.869384835 | 59.46 | 0.858617 | 0.853622474 | 55.95 |
model_type | roc_test | accuracy_test | ks_test | roc_oot1 | accuracy_oot1 |
|---|---|---|---|---|---|
neuralNetwork | 0.813792 | 0.849134 | 46.58 | 0.856175 | 0.8582 |
randomForest | 0.806239 | 0.840979 | 44.3 | 0.834677 | 0.8575 |
logistic | 0.719284 | 0.805301 | 32.19 | 0.751941 | 0.8106 |
gradientBoosting | 0.816005 | 0.850153 | 47.22 | 0.870804 | 0.8643 |
model_type | ks_oot1 | roc_oot2 | accuracy_oot2 | ks_oot2 | selected_model |
|---|---|---|---|---|---|
neuralNetwork | 54.43 | 0.856175108 | 0.8582 | 54.43 | Champion |
randomForest | 49.99 | 0.834676561 | 0.8575 | 49.99 | Challenger |
logistic | 36.49 | 0.751941185 | 0.8106 | 36.49 | |
gradientBoosting | 57.51 | 0.870803793 | 0.8643 | 57.51 |
Kolmogorov-Smirnov Statistic
- |
Neural Network Test
- |
This module is intended to help you create quick, automated experiments and is in no way meant to replace the model-building activity.
Question: We challenge you to build a separate pipeline or integrate code flow to accommodate continuous targets.
Summary
We learned the challenges of model management and deployment.
We now know how to use MLflow for managing experiments and deploying models.
We explored how to build custom pipelines for various model-building activities.
We saw how pipelines can be stacked together to create an automated pipeline.
Great job! You are now familiar with some of the key concepts that will be useful in putting a model into production. This should give you a fair idea of how you want to manage a model lifecycle as you are building your data-preparation pipelines. In the next chapter, we will cover some of the tips, tricks, and interesting topics that can be useful in your day-to-day work.



