Much of the programming work in data analysis and modeling is spent on data preparation: loading, cleaning, transforming, and rearranging. Sometimes the way that data is stored in files or databases is not the way you need it for a data processing application. Many people choose to do ad hoc processing of data from one form to another using a general purpose programming, like Python, Perl, R, or Java, or UNIX text processing tools like sed or awk. Fortunately, pandas along with the Python standard library provide you with a high-level, flexible, and high-performance set of core manipulations and algorithms to enable you to wrangle data into the right form without much trouble.
If you identify a type of data manipulation that isn’t anywhere in this book or elsewhere in the pandas library, feel free to suggest it on the mailing list or GitHub site. Indeed, much of the design and implementation of pandas has been driven by the needs of real world applications.
Data contained in pandas objects can be combined together in a number of built-in ways:
pandas.mergeconnects rows in DataFrames based on one or more keys. This will be familiar to users of SQL or other relational databases, as it implements database join operations.pandas.concatglues or stacks together objects along an axis.combine_firstinstance method enables splicing together overlapping data to fill in missing values in one object with values from another.
I will address each of these and give a number of examples. They’ll be utilized in examples throughout the rest of the book.
Merge or join
operations combine data sets by linking rows using one or more
keys. These operations are central to relational
databases. The merge function in
pandas is the main entry point for using these algorithms on your
data.
Let’s start with a simple example:
In [15]: df1 = DataFrame({'key': ['b', 'b', 'a', 'c', 'a', 'a', 'b'],
....: 'data1': range(7)})
In [16]: df2 = DataFrame({'key': ['a', 'b', 'd'],
....: 'data2': range(3)})
In [17]: df1 In [18]: df2
Out[17]: Out[18]:
data1 key data2 key
0 0 b 0 0 a
1 1 b 1 1 b
2 2 a 2 2 d
3 3 c
4 4 a
5 5 a
6 6 b This is an example of a many-to-one merge situation; the data
in df1 has multiple rows labeled
a and b, whereas df2 has only one row for each value in the
key column. Calling merge with these objects we obtain:
In [19]: pd.merge(df1, df2) Out[19]: data1 key data2 0 0 b 1 1 1 b 1 2 6 b 1 3 2 a 0 4 4 a 0 5 5 a 0
Note that I didn’t specify which column to join on. If not
specified, merge uses the overlapping
column names as the keys. It’s a good practice to specify explicitly,
though:
In [20]: pd.merge(df1, df2, on='key') Out[20]: data1 key data2 0 0 b 1 1 1 b 1 2 6 b 1 3 2 a 0 4 4 a 0 5 5 a 0
If the column names are different in each object, you can specify them separately:
In [21]: df3 = DataFrame({'lkey': ['b', 'b', 'a', 'c', 'a', 'a', 'b'],
....: 'data1': range(7)})
In [22]: df4 = DataFrame({'rkey': ['a', 'b', 'd'],
....: 'data2': range(3)})
In [23]: pd.merge(df3, df4, left_on='lkey', right_on='rkey')
Out[23]:
data1 lkey data2 rkey
0 0 b 1 b
1 1 b 1 b
2 6 b 1 b
3 2 a 0 a
4 4 a 0 a
5 5 a 0 aYou probably noticed that the 'c' and 'd'
values and associated data are missing from the result. By default
merge does an
'inner' join; the keys in the result are the
intersection. Other possible options are 'left',
'right', and 'outer'. The outer join takes the union of the
keys, combining the effect of applying both left and right joins:
In [24]: pd.merge(df1, df2, how='outer') Out[24]: data1 key data2 0 0 b 1 1 1 b 1 2 6 b 1 3 2 a 0 4 4 a 0 5 5 a 0 6 3 c NaN 7 NaN d 2
Many-to-many merges have well-defined though not necessarily intuitive behavior. Here’s an example:
In [25]: df1 = DataFrame({'key': ['b', 'b', 'a', 'c', 'a', 'b'],
....: 'data1': range(6)})
In [26]: df2 = DataFrame({'key': ['a', 'b', 'a', 'b', 'd'],
....: 'data2': range(5)})
In [27]: df1 In [28]: df2
Out[27]: Out[28]:
data1 key data2 key
0 0 b 0 0 a
1 1 b 1 1 b
2 2 a 2 2 a
3 3 c 3 3 b
4 4 a 4 4 d
5 5 b
In [29]: pd.merge(df1, df2, on='key', how='left')
Out[29]:
data1 key data2
0 0 b 1
1 0 b 3
2 1 b 1
3 1 b 3
4 5 b 1
.. ... .. ...
6 2 a 0
7 2 a 2
8 4 a 0
9 4 a 2
10 3 c NaN
[11 rows x 3 columns]Many-to-many joins form the Cartesian product of the rows. Since
there were 3 'b' rows in the left
DataFrame and 2 in the right one, there are 6 'b' rows in the result. The join method only
affects the distinct key values appearing in the result:
In [30]: pd.merge(df1, df2, how='inner') Out[30]: data1 key data2 0 0 b 1 1 0 b 3 2 1 b 1 3 1 b 3 4 5 b 1 5 5 b 3 6 2 a 0 7 2 a 2 8 4 a 0 9 4 a 2
To merge with multiple keys, pass a list of column names:
In [31]: left = DataFrame({'key1': ['foo', 'foo', 'bar'],
....: 'key2': ['one', 'two', 'one'],
....: 'lval': [1, 2, 3]})
In [32]: right = DataFrame({'key1': ['foo', 'foo', 'bar', 'bar'],
....: 'key2': ['one', 'one', 'one', 'two'],
....: 'rval': [4, 5, 6, 7]})
In [33]: pd.merge(left, right, on=['key1', 'key2'], how='outer')
Out[33]:
key1 key2 lval rval
0 foo one 1 4
1 foo one 1 5
2 foo two 2 NaN
3 bar one 3 6
4 bar two NaN 7To determine which key combinations will appear in the result depending on the choice of merge method, think of the multiple keys as forming an array of tuples to be used as a single join key (even though it’s not actually implemented that way).
Caution
When joining columns-on-columns, the indexes on the passed DataFrame objects are discarded.
A last issue to consider in merge operations is the treatment of
overlapping column names. While you can address the overlap manually
(see the later section on renaming axis labels), merge has a suffixes option for specifying strings to
append to overlapping names in the left and right DataFrame
objects:
In [34]: pd.merge(left, right, on='key1')
Out[34]:
key1 key2_x lval key2_y rval
0 foo one 1 one 4
1 foo one 1 one 5
2 foo two 2 one 4
3 foo two 2 one 5
4 bar one 3 one 6
5 bar one 3 two 7
In [35]: pd.merge(left, right, on='key1', suffixes=('_left', '_right'))
Out[35]:
key1 key2_left lval key2_right rval
0 foo one 1 one 4
1 foo one 1 one 5
2 foo two 2 one 4
3 foo two 2 one 5
4 bar one 3 one 6
5 bar one 3 two 7See Table 7-1 for an argument
reference on merge. Joining on index
is the subject of the next section.
Table 7-1. merge function arguments
In some cases, the merge key or keys in a DataFrame will
be found in its index. In this case, you can pass left_index=True or right_index=True (or both) to indicate that
the index should be used as the merge key:
In [36]: left1 = DataFrame({'key': ['a', 'b', 'a', 'a', 'b', 'c'],
....: 'value': range(6)})
In [37]: right1 = DataFrame({'group_val': [3.5, 7]}, index=['a', 'b'])
In [38]: left1 In [39]: right1
Out[38]: Out[39]:
key value group_val
0 a 0 a 3.5
1 b 1 b 7.0
2 a 2
3 a 3
4 b 4
5 c 5
In [40]: pd.merge(left1, right1, left_on='key', right_index=True)
Out[40]:
key value group_val
0 a 0 3.5
2 a 2 3.5
3 a 3 3.5
1 b 1 7.0
4 b 4 7.0Since the default merge method is to intersect the join keys, you can instead form the union of them with an outer join:
In [41]: pd.merge(left1, right1, left_on='key', right_index=True, how='outer') Out[41]: key value group_val 0 a 0 3.5 2 a 2 3.5 3 a 3 3.5 1 b 1 7.0 4 b 4 7.0 5 c 5 NaN
With hierarchically-indexed data, things are a bit more complicated:
In [42]: lefth = DataFrame({'key1': ['Ohio', 'Ohio', 'Ohio', 'Nevada', 'Nevada'],
....: 'key2': [2000, 2001, 2002, 2001, 2002],
....: 'data': np.arange(5.)})
In [43]: righth = DataFrame(np.arange(12).reshape((6, 2)),
....: index=[['Nevada', 'Nevada', 'Ohio', 'Ohio', 'Ohio', 'Ohio'],
....: [2001, 2000, 2000, 2000, 2001, 2002]],
....: columns=['event1', 'event2'])
In [44]: lefth In [45]: righth
Out[44]: Out[45]:
data key1 key2 event1 event2
0 0 Ohio 2000 Nevada 2001 0 1
1 1 Ohio 2001 2000 2 3
2 2 Ohio 2002 Ohio 2000 4 5
3 3 Nevada 2001 2000 6 7
4 4 Nevada 2002 2001 8 9
2002 10 11In this case, you have to indicate multiple columns to merge on as a list (pay attention to the handling of duplicate index values):
In [46]: pd.merge(lefth, righth, left_on=['key1', 'key2'], right_index=True) Out[46]: data key1 key2 event1 event2 0 0 Ohio 2000 4 5 0 0 Ohio 2000 6 7 1 1 Ohio 2001 8 9 2 2 Ohio 2002 10 11 3 3 Nevada 2001 0 1 In [47]: pd.merge(lefth, righth, left_on=['key1', 'key2'], ....: right_index=True, how='outer') Out[47]: data key1 key2 event1 event2 0 0 Ohio 2000 4 5 0 0 Ohio 2000 6 7 1 1 Ohio 2001 8 9 2 2 Ohio 2002 10 11 3 3 Nevada 2001 0 1 4 4 Nevada 2002 NaN NaN 4 NaN Nevada 2000 2 3
Using the indexes of both sides of the merge is also not an issue:
In [48]: left2 = DataFrame([[1., 2.], [3., 4.], [5., 6.]], index=['a', 'c', 'e'],
....: columns=['Ohio', 'Nevada'])
In [49]: right2 = DataFrame([[7., 8.], [9., 10.], [11., 12.], [13, 14]],
....: index=['b', 'c', 'd', 'e'], columns=['Missouri', 'Alabama'])
In [50]: left2 In [51]: right2
Out[50]: Out[51]:
Ohio Nevada Missouri Alabama
a 1 2 b 7 8
c 3 4 c 9 10
e 5 6 d 11 12
e 13 14
In [52]: pd.merge(left2, right2, how='outer', left_index=True, right_index=True)
Out[52]:
Ohio Nevada Missouri Alabama
a 1 2 NaN NaN
b NaN NaN 7 8
c 3 4 9 10
d NaN NaN 11 12
e 5 6 13 14DataFrame has a more convenient join instance for
merging by index. It can also be used to combine together many DataFrame
objects having the same or similar indexes but non-overlapping columns.
In the prior example, we could have written:
In [53]: left2.join(right2, how='outer') Out[53]: Ohio Nevada Missouri Alabama a 1 2 NaN NaN b NaN NaN 7 8 c 3 4 9 10 d NaN NaN 11 12 e 5 6 13 14
In part for legacy reasons (much earlier versions of pandas),
DataFrame’s join method performs a
left join on the join keys. It also supports joining the index of the
passed DataFrame on one of the columns of the calling DataFrame:
In [54]: left1.join(right1, on='key') Out[54]: key value group_val 0 a 0 3.5 1 b 1 7.0 2 a 2 3.5 3 a 3 3.5 4 b 4 7.0 5 c 5 NaN
Lastly, for simple index-on-index merges, you can pass a list of
DataFrames to join as an alternative
to using the more general concat function
described below:
In [55]: another = DataFrame([[7., 8.], [9., 10.], [11., 12.], [16., 17.]], ....: index=['a', 'c', 'e', 'f'], columns=['New York', 'Oregon']) In [56]: left2.join([right2, another]) Out[56]: Ohio Nevada Missouri Alabama New York Oregon a 1 2 NaN NaN 7 8 c 3 4 9 10 9 10 e 5 6 13 14 11 12 In [57]: left2.join([right2, another], how='outer') Out[57]: Ohio Nevada Missouri Alabama New York Oregon a 1 2 NaN NaN 7 8 b NaN NaN 7 8 NaN NaN c 3 4 9 10 9 10 d NaN NaN 11 12 NaN NaN e 5 6 13 14 11 12 f NaN NaN NaN NaN 16 17
Another kind of data combination operation is
alternatively referred to as concatenation, binding, or stacking. NumPy
has a concatenate function for doing
this with raw NumPy arrays:
In [58]: arr = np.arange(12).reshape((3, 4))
In [59]: arr
Out[59]:
array([[ 0, 1, 2, 3],
[ 4, 5, 6, 7],
[ 8, 9, 10, 11]])
In [60]: np.concatenate([arr, arr], axis=1)
Out[60]:
array([[ 0, 1, 2, 3, 0, 1, 2, 3],
[ 4, 5, 6, 7, 4, 5, 6, 7],
[ 8, 9, 10, 11, 8, 9, 10, 11]])In the context of pandas objects such as Series and DataFrame, having labeled axes enable you to further generalize array concatenation. In particular, you have a number of additional things to think about:
If the objects are indexed differently on the other axes, should the collection of axes be unioned or intersected?
Do the groups need to be identifiable in the resulting object?
Does the concatenation axis matter at all?
The concat function in pandas provides a
consistent way to address each of these concerns. I’ll give a number of
examples to illustrate how it works. Suppose we have three Series with
no index overlap:
In [61]: s1 = Series([0, 1], index=['a', 'b']) In [62]: s2 = Series([2, 3, 4], index=['c', 'd', 'e']) In [63]: s3 = Series([5, 6], index=['f', 'g'])
Calling concat with these
object in a list glues together the values and indexes:
In [64]: pd.concat([s1, s2, s3]) Out[64]: a 0 b 1 c 2 d 3 e 4 f 5 g 6 dtype: int64
By default concat works along
axis=0, producing another Series. If
you pass axis=1, the result will
instead be a DataFrame (axis=1 is the
columns):
In [65]: pd.concat([s1, s2, s3], axis=1)
Out[65]:
0 1 2
a 0 NaN NaN
b 1 NaN NaN
c NaN 2 NaN
d NaN 3 NaN
e NaN 4 NaN
f NaN NaN 5
g NaN NaN 6In this case there is no overlap on the other axis, which as you
can see is the sorted union (the 'outer' join) of the indexes. You can instead
intersect them by passing join='inner':
In [66]: s4 = pd.concat([s1 * 5, s3])
In [67]: pd.concat([s1, s4], axis=1) In [68]: pd.concat([s1, s4], axis=1, join='inner')
Out[67]: Out[68]:
0 1 0 1
a 0 0 a 0 0
b 1 5 b 1 5
f NaN 5
g NaN 6You can even specify the axes to be used on the other axes with
join_axes:
In [69]: pd.concat([s1, s4], axis=1, join_axes=[['a', 'c', 'b', 'e']])
Out[69]:
0 1
a 0 0
c NaN NaN
b 1 5
e NaN NaNOne issue is that the concatenated pieces are not identifiable in
the result. Suppose instead you wanted to create a hierarchical index on
the concatenation axis. To do this, use the keys argument:
In [70]: result = pd.concat([s1, s1, s3], keys=['one', 'two', 'three'])
In [71]: result
Out[71]:
one a 0
b 1
two a 0
b 1
three f 5
g 6
dtype: int64
# Much more on the unstack function later
In [72]: result.unstack()
Out[72]:
a b f g
one 0 1 NaN NaN
two 0 1 NaN NaN
three NaN NaN 5 6In the case of combining Series along axis=1, the keys become the DataFrame column
headers:
In [73]: pd.concat([s1, s2, s3], axis=1, keys=['one', 'two', 'three']) Out[73]: one two three a 0 NaN NaN b 1 NaN NaN c NaN 2 NaN d NaN 3 NaN e NaN 4 NaN f NaN NaN 5 g NaN NaN 6
The same logic extends to DataFrame objects:
In [74]: df1 = DataFrame(np.arange(6).reshape(3, 2), index=['a', 'b', 'c'],
....: columns=['one', 'two'])
In [75]: df2 = DataFrame(5 + np.arange(4).reshape(2, 2), index=['a', 'c'],
....: columns=['three', 'four'])
In [76]: pd.concat([df1, df2], axis=1, keys=['level1', 'level2'])
Out[76]:
level1 level2
one two three four
a 0 1 5 6
b 2 3 NaN NaN
c 4 5 7 8If you pass a dict of objects instead of a list, the dict’s keys
will be used for the keys
option:
In [77]: pd.concat({'level1': df1, 'level2': df2}, axis=1)
Out[77]:
level1 level2
one two three four
a 0 1 5 6
b 2 3 NaN NaN
c 4 5 7 8There are a couple of additional arguments governing how the hierarchical index is created (see Table 7-2):
In [78]: pd.concat([df1, df2], axis=1, keys=['level1', 'level2'], ....: names=['upper', 'lower']) Out[78]: upper level1 level2 lower one two three four a 0 1 5 6 b 2 3 NaN NaN c 4 5 7 8
A last consideration concerns DataFrames in which the row index is not meaningful in the context of the analysis:
In [79]: df1 = DataFrame(np.random.randn(3, 4), columns=['a', 'b', 'c', 'd'])
In [80]: df2 = DataFrame(np.random.randn(2, 3), columns=['b', 'd', 'a'])
In [81]: df1 In [82]: df2
Out[81]: Out[82]:
a b c d b d a
0 -0.204708 0.478943 -0.519439 -0.555730 0 0.274992 0.228913 1.352917
1 1.965781 1.393406 0.092908 0.281746 1 0.886429 -2.001637 -0.371843
2 0.769023 1.246435 1.007189 -1.296221In this case, you can pass ignore_index=True:
In [83]: pd.concat([df1, df2], ignore_index=True)
Out[83]:
a b c d
0 -0.204708 0.478943 -0.519439 -0.555730
1 1.965781 1.393406 0.092908 0.281746
2 0.769023 1.246435 1.007189 -1.296221
3 1.352917 0.274992 NaN 0.228913
4 -0.371843 0.886429 NaN -2.001637Table 7-2. concat function arguments
Another data combination situation can’t be expressed as
either a merge or concatenation operation. You may have two datasets
whose indexes overlap in full or part. As a motivating example, consider
NumPy’s where function, which
expressed a vectorized if-else:
In [84]: a = Series([np.nan, 2.5, np.nan, 3.5, 4.5, np.nan],
....: index=['f', 'e', 'd', 'c', 'b', 'a'])
In [85]: b = Series(np.arange(len(a), dtype=np.float64),
....: index=['f', 'e', 'd', 'c', 'b', 'a'])
In [86]: b[-1] = np.nan
In [87]: a In [88]: b In [89]: np.where(pd.isnull(a), b, a)
Out[87]: Out[88]: Out[89]: array([ 0. , 2.5, 2. ,
3.5, 4.5, nan])
f NaN f 0
e 2.5 e 1
d NaN d 2
c 3.5 c 3
b 4.5 b 4
a NaN a NaN
dtype: float64 dtype: float64Series has a combine_first method,
which performs the equivalent of this operation plus data
alignment:
In [90]: b[:-2].combine_first(a[2:]) Out[90]: a NaN b 4.5 c 3.0 d 2.0 e 1.0 f 0.0 dtype: float64
With DataFrames, combine_first naturally
does the same thing column by column, so you can think of it as
“patching” missing data in the calling object with data from the object
you pass:
In [91]: df1 = DataFrame({'a': [1., np.nan, 5., np.nan],
....: 'b': [np.nan, 2., np.nan, 6.],
....: 'c': range(2, 18, 4)})
In [92]: df2 = DataFrame({'a': [5., 4., np.nan, 3., 7.],
....: 'b': [np.nan, 3., 4., 6., 8.]})
In [93]: df1.combine_first(df2)
Out[93]:
a b c
0 1 NaN 2
1 4 2 6
2 5 4 10
3 3 6 14
4 7 8 NaNThere are a number of fundamental operations for rearranging tabular data. These are alternatingly referred to as reshape or pivot operations.
Hierarchical indexing provides a consistent way to rearrange data in a DataFrame. There are two primary actions:
stack: this “rotates” or pivots from the columns in the data to the rowsunstack: this pivots from the rows into the columns
I’ll illustrate these operations through a series of examples. Consider a small DataFrame with string arrays as row and column indexes:
In [94]: data = DataFrame(np.arange(6).reshape((2, 3)), ....: index=pd.Index(['Ohio', 'Colorado'], name='state'), ....: columns=pd.Index(['one', 'two', 'three'], name='number')) In [95]: data Out[95]: number one two three state Ohio 0 1 2 Colorado 3 4 5
Using the stack method on this
data pivots the columns into the rows, producing a Series:
In [96]: result = data.stack()
In [97]: result
Out[97]:
state number
Ohio one 0
two 1
three 2
Colorado one 3
two 4
three 5
dtype: int64From a hierarchically-indexed Series, you can rearrange the data
back into a DataFrame with unstack:
In [98]: result.unstack() Out[98]: number one two three state Ohio 0 1 2 Colorado 3 4 5
By default the innermost level is unstacked (same with stack). You can unstack a different level by
passing a level number or name:
In [99]: result.unstack(0) In [100]: result.unstack('state')
Out[99]: Out[100]:
state Ohio Colorado state Ohio Colorado
number number
one 0 3 one 0 3
two 1 4 two 1 4
three 2 5 three 2 5 Unstacking might introduce missing data if all of the values in the level aren’t found in each of the subgroups:
In [101]: s1 = Series([0, 1, 2, 3], index=['a', 'b', 'c', 'd'])
In [102]: s2 = Series([4, 5, 6], index=['c', 'd', 'e'])
In [103]: data2 = pd.concat([s1, s2], keys=['one', 'two'])
In [104]: data2.unstack()
Out[104]:
a b c d e
one 0 1 2 3 NaN
two NaN NaN 4 5 6Stacking filters out missing data by default, so the operation is easily invertible:
In [105]: data2.unstack().stack() In [106]: data2.unstack().stack(dropna=False)
Out[105]: Out[106]:
one a 0 one a 0
b 1 b 1
c 2 c 2
d 3 d 3
two c 4 e NaN
d 5 two a NaN
e 6 b NaN
dtype: float64 c 4
d 5
e 6
dtype: float64When unstacking in a DataFrame, the level unstacked becomes the lowest level in the result:
In [107]: df = DataFrame({'left': result, 'right': result + 5},
.....: columns=pd.Index(['left', 'right'], name='side'))
In [108]: df
Out[108]:
side left right
state number
Ohio one 0 5
two 1 6
three 2 7
Colorado one 3 8
two 4 9
three 5 10
In [109]: df.unstack('state') In [110]: df.unstack('state').stack('side')
Out[109]: Out[110]:
side left right state Ohio Colorado
state Ohio Colorado Ohio Colorado number side
number one left 0 3
one 0 3 5 8 right 5 8
two 1 4 6 9 two left 1 4
three 2 5 7 10 right 6 9
three left 2 5
right 7 10A common way to store multiple time series in databases and CSV is in so-called long or stacked format:
data = pd.read_csv('ch07/macrodata.csv')
periods = pd.PeriodIndex(year=data.year, quarter=data.quarter, name='date')
data = DataFrame(data.to_records(),
columns=pd.Index(['realgdp', 'infl', 'unemp'], name='item'),
index=periods.to_timestamp('D', 'end'))
ldata = data.stack().reset_index().rename(columns={0: 'value'})
In [116]: ldata[:10]
Out[116]:
date item value
0 1959-03-31 realgdp 2710.349
1 1959-03-31 infl 0.000
2 1959-03-31 unemp 5.800
3 1959-06-30 realgdp 2778.801
4 1959-06-30 infl 2.340
5 1959-06-30 unemp 5.100
6 1959-09-30 realgdp 2775.488
7 1959-09-30 infl 2.740
8 1959-09-30 unemp 5.300
9 1959-12-31 realgdp 2785.204Data is frequently stored this way in relational databases like
MySQL as a fixed schema (column names and data types) allows the number
of distinct values in the item column
to increase or decrease as data is added or deleted in the table. In the
above example date and item would usually be the primary keys (in
relational database parlance), offering both relational integrity and
easier joins and programmatic queries in many cases. The downside, of
course, is that the data may not be easy to work with in long format;
you might prefer to have a DataFrame containing one column per distinct
item value indexed by timestamps in
the date column. DataFrame’s pivot method performs exactly this
transformation:
In [117]: pivoted = ldata.pivot('date', 'item', 'value')
In [118]: pivoted.head()
Out[118]:
item infl realgdp unemp
date
1959-03-31 0.00 2710.349 5.8
1959-06-30 2.34 2778.801 5.1
1959-09-30 2.74 2775.488 5.3
1959-12-31 0.27 2785.204 5.6
1960-03-31 2.31 2847.699 5.2The first two values passed are the columns to be used as the row and column index, and finally an optional value column to fill the DataFrame. Suppose you had two value columns that you wanted to reshape simultaneously:
In [119]: ldata['value2'] = np.random.randn(len(ldata))
In [120]: ldata[:10]
Out[120]:
date item value value2
0 1959-03-31 realgdp 2710.349 1.669025
1 1959-03-31 infl 0.000 -0.438570
2 1959-03-31 unemp 5.800 -0.539741
3 1959-06-30 realgdp 2778.801 0.476985
4 1959-06-30 infl 2.340 3.248944
5 1959-06-30 unemp 5.100 -1.021228
6 1959-09-30 realgdp 2775.488 -0.577087
7 1959-09-30 infl 2.740 0.124121
8 1959-09-30 unemp 5.300 0.302614
9 1959-12-31 realgdp 2785.204 0.523772By omitting the last argument, you obtain a DataFrame with hierarchical columns:
In [121]: pivoted = ldata.pivot('date', 'item')
In [122]: pivoted[:5]
Out[122]:
value value2
item infl realgdp unemp infl realgdp unemp
date
1959-03-31 0.00 2710.349 5.8 -0.438570 1.669025 -0.539741
1959-06-30 2.34 2778.801 5.1 3.248944 0.476985 -1.021228
1959-09-30 2.74 2775.488 5.3 0.124121 -0.577087 0.302614
1959-12-31 0.27 2785.204 5.6 0.000940 0.523772 1.343810
1960-03-31 2.31 2847.699 5.2 -0.831154 -0.713544 -2.370232
In [123]: pivoted['value'][:5]
Out[123]:
item infl realgdp unemp
date
1959-03-31 0.00 2710.349 5.8
1959-06-30 2.34 2778.801 5.1
1959-09-30 2.74 2775.488 5.3
1959-12-31 0.27 2785.204 5.6
1960-03-31 2.31 2847.699 5.2Note that pivot is just a
shortcut for creating a hierarchical index using set_index and reshaping
with unstack:
In [124]: unstacked = ldata.set_index(['date', 'item']).unstack('item')
In [125]: unstacked[:7]
Out[125]:
value value2
item infl realgdp unemp infl realgdp unemp
date
1959-03-31 0.00 2710.349 5.8 -0.438570 1.669025 -0.539741
1959-06-30 2.34 2778.801 5.1 3.248944 0.476985 -1.021228
1959-09-30 2.74 2775.488 5.3 0.124121 -0.577087 0.302614
1959-12-31 0.27 2785.204 5.6 0.000940 0.523772 1.343810
1960-03-31 2.31 2847.699 5.2 -0.831154 -0.713544 -2.370232
1960-06-30 0.14 2834.390 5.2 -0.860757 -1.860761 0.560145
1960-09-30 2.70 2839.022 5.6 0.119827 -1.265934 -1.063512So far in this chapter we’ve been concerned with rearranging data. Filtering, cleaning, and other tranformations are another class of important operations.
Duplicate rows may be found in a DataFrame for any number of reasons. Here is an example:
In [126]: data = DataFrame({'k1': ['one'] * 3 + ['two'] * 4,
.....: 'k2': [1, 1, 2, 3, 3, 4, 4]})
In [127]: data
Out[127]:
k1 k2
0 one 1
1 one 1
2 one 2
3 two 3
4 two 3
5 two 4
6 two 4The DataFrame method duplicated returns a
boolean Series indicating whether each row is a duplicate or not:
In [128]: data.duplicated() Out[128]: 0 False 1 True 2 False 3 False 4 True 5 False 6 True dtype: bool
Relatedly, drop_duplicates returns
a DataFrame where the duplicated array is
False:
In [129]: data.drop_duplicates()
Out[129]:
k1 k2
0 one 1
2 one 2
3 two 3
5 two 4Both of these methods by default consider all of the columns;
alternatively you can specify any subset of them to detect duplicates.
Suppose we had an additional column of values and wanted to filter
duplicates only based on the 'k1'
column:
In [130]: data['v1'] = range(7)
In [131]: data.drop_duplicates(['k1'])
Out[131]:
k1 k2 v1
0 one 1 0
3 two 3 3duplicated and drop_duplicates by default keep the first
observed value combination. Passing take_last=True will return the last
one:
In [132]: data.drop_duplicates(['k1', 'k2'], take_last=True)
Out[132]:
k1 k2 v1
1 one 1 1
2 one 2 2
4 two 3 4
6 two 4 6For many data sets, you may wish to perform some transformation based on the values in an array, Series, or column in a DataFrame. Consider the following hypothetical data collected about some kinds of meat:
In [133]: data = DataFrame({'food': ['bacon', 'pulled pork', 'bacon', 'Pastrami',
.....: 'corned beef', 'Bacon', 'pastrami', 'honey ham',
.....: 'nova lox'],
.....: 'ounces': [4, 3, 12, 6, 7.5, 8, 3, 5, 6]})
In [134]: data
Out[134]:
food ounces
0 bacon 4.0
1 pulled pork 3.0
2 bacon 12.0
3 Pastrami 6.0
4 corned beef 7.5
5 Bacon 8.0
6 pastrami 3.0
7 honey ham 5.0
8 nova lox 6.0Suppose you wanted to add a column indicating the type of animal that each food came from. Let’s write down a mapping of each distinct meat type to the kind of animal:
meat_to_animal = {
'bacon': 'pig',
'pulled pork': 'pig',
'pastrami': 'cow',
'corned beef': 'cow',
'honey ham': 'pig',
'nova lox': 'salmon'
}The map method on a Series
accepts a function or dict-like object containing a mapping, but here we
have a small problem in that some of the meats above are capitalized and
others are not. Thus, we also need to convert each value to lower
case:
In [136]: data['animal'] = data['food'].map(str.lower).map(meat_to_animal)
In [137]: data
Out[137]:
food ounces animal
0 bacon 4.0 pig
1 pulled pork 3.0 pig
2 bacon 12.0 pig
3 Pastrami 6.0 cow
4 corned beef 7.5 cow
5 Bacon 8.0 pig
6 pastrami 3.0 cow
7 honey ham 5.0 pig
8 nova lox 6.0 salmonWe could also have passed a function that does all the work:
In [138]: data['food'].map(lambda x: meat_to_animal[x.lower()]) Out[138]: 0 pig 1 pig 2 pig 3 cow 4 cow 5 pig 6 cow 7 pig 8 salmon Name: food, dtype: object
Using map is a convenient way
to perform element-wise transformations and other data cleaning-related
operations.
Filling in missing data with the fillna method can be
thought of as a special case of more general value replacement. While
map, as you’ve seen
above, can be used to modify a subset of values in an object, replace provides a
simpler and more flexible way to do so. Let’s consider this
Series:
In [139]: data = Series([1., -999., 2., -999., -1000., 3.]) In [140]: data Out[140]: 0 1 1 -999 2 2 3 -999 4 -1000 5 3 dtype: float64
The -999 values might be
sentinel values for missing data. To replace these with NA values that
pandas understands, we can use replace, producing a new Series:
In [141]: data.replace(-999, np.nan) Out[141]: 0 1 1 NaN 2 2 3 NaN 4 -1000 5 3 dtype: float64
If you want to replace multiple values at once, you instead pass a list then the substitute value:
In [142]: data.replace([-999, -1000], np.nan) Out[142]: 0 1 1 NaN 2 2 3 NaN 4 NaN 5 3 dtype: float64
To use a different replacement for each value, pass a list of substitutes:
In [143]: data.replace([-999, -1000], [np.nan, 0]) Out[143]: 0 1 1 NaN 2 2 3 NaN 4 0 5 3 dtype: float64
The argument passed can also be a dict:
In [144]: data.replace({-999: np.nan, -1000: 0})
Out[144]:
0 1
1 NaN
2 2
3 NaN
4 0
5 3
dtype: float64Like values in a Series, axis labels can be similarly transformed by a function or mapping of some form to produce new, differently labeled objects. The axes can also be modified in place without creating a new data structure. Here’s a simple example:
In [145]: data = DataFrame(np.arange(12).reshape((3, 4)), .....: index=['Ohio', 'Colorado', 'New York'], .....: columns=['one', 'two', 'three', 'four'])
Like a Series, the axis indexes have a map method:
In [146]: data.index.map(str.upper) Out[146]: array(['OHIO', 'COLORADO', 'NEW YORK'], dtype=object)
You can assign to index,
modifying the DataFrame in place:
In [147]: data.index = data.index.map(str.upper)
In [148]: data
Out[148]:
one two three four
OHIO 0 1 2 3
COLORADO 4 5 6 7
NEW YORK 8 9 10 11If you want to create a transformed version of a data set without
modifying the original, a useful method is rename:
In [149]: data.rename(index=str.title, columns=str.upper)
Out[149]:
ONE TWO THREE FOUR
Ohio 0 1 2 3
Colorado 4 5 6 7
New York 8 9 10 11Notably, rename can be used in
conjunction with a dict-like object providing new values for a subset of
the axis labels:
In [150]: data.rename(index={'OHIO': 'INDIANA'},
.....: columns={'three': 'peekaboo'})
Out[150]:
one two peekaboo four
INDIANA 0 1 2 3
COLORADO 4 5 6 7
NEW YORK 8 9 10 11rename saves having to
copy the DataFrame manually and assign to its index and columns attributes. Should you wish to modify
a data set in place, pass inplace=True:
# Always returns a reference to a DataFrame
In [151]: _ = data.rename(index={'OHIO': 'INDIANA'}, inplace=True)
In [152]: data
Out[152]:
one two three four
INDIANA 0 1 2 3
COLORADO 4 5 6 7
NEW YORK 8 9 10 11Continuous data is often discretized or otherwised separated into “bins” for analysis. Suppose you have data about a group of people in a study, and you want to group them into discrete age buckets:
In [153]: ages = [20, 22, 25, 27, 21, 23, 37, 31, 61, 45, 41, 32]
Let’s divide these into bins of 18 to 25, 26 to 35, 36 to 60, and
finally 61 and older. To do so, you have to use cut, a function in
pandas:
In [154]: bins = [18, 25, 35, 60, 100] In [155]: cats = pd.cut(ages, bins) In [156]: cats Out[156]: (18, 25] (18, 25] (18, 25] ... (35, 60] (35, 60] (25, 35] Levels (4): Index(['(18, 25]', '(25, 35]', '(35, 60]', '(60, 100]'], dtype=object) Length: 12
The object pandas returns is a special Categorical object. You
can treat it like an array of strings indicating the bin name;
internally it contains a levels array
indicating the distinct category names along with a labeling for the
ages data in the labels attribute:
In [157]: cats.labels Out[157]: array([0, 0, 0, 1, 0, 0, 2, 1, 3, 2, 2, 1]) In [158]: cats.levels Out[158]: Index([u'(18, 25]', u'(25, 35]', u'(35, 60]', u'(60, 100]'], dtype='object') In [159]: pd.value_counts(cats) Out[159]: (18, 25] 5 (35, 60] 3 (25, 35] 3 (60, 100] 1 dtype: int64
Consistent with mathematical notation for intervals, a parenthesis
means that the side is open while the square
bracket means it is closed (inclusive). Which side
is closed can be changed by passing right=False:
In [160]: pd.cut(ages, [18, 26, 36, 61, 100], right=False) Out[160]: [18, 26) [18, 26) [18, 26) ... [36, 61) [36, 61) [26, 36) Levels (4): Index(['[18, 26)', '[26, 36)', '[36, 61)', '[61, 100)'], dtype=object) Length: 12
You can also pass your own bin names by passing a list or array to
the labels option:
In [161]: group_names = ['Youth', 'YoungAdult', 'MiddleAged', 'Senior'] In [162]: pd.cut(ages, bins, labels=group_names) Out[162]: Youth Youth Youth ... MiddleAged MiddleAged YoungAdult Levels (4): Index(['Youth', 'YoungAdult', 'MiddleAged', 'Senior'], dtype=object) Length: 12
If you pass cut a integer number of
bins instead of explicit bin edges, it will compute equal-length bins
based on the minimum and maximum values in the data. Consider the case
of some uniformly distributed data chopped into fourths:
In [163]: data = np.random.rand(20)
In [164]: pd.cut(data, 4, precision=2)
Out[164]:
(0.45, 0.67]
(0.23, 0.45]
(0.0037, 0.23]
...
(0.0037, 0.23]
(0.23, 0.45]
(0.23, 0.45]
Levels (4): Index(['(0.0037, 0.23]', '(0.23, 0.45]', '(0.45, 0.67]',
'(0.67, 0.9]'], dtype=object)
Length: 20A closely related function, qcut, bins the data
based on sample quantiles. Depending on the distribution of the data,
using cut will not usually
result in each bin having the same number of data points. Since
qcut uses sample
quantiles instead, by definition you will obtain roughly equal-size
bins:
In [165]: data = np.random.randn(1000) # Normally distributed
In [166]: cats = pd.qcut(data, 4) # Cut into quartiles
In [167]: cats
Out[167]:
(-0.022, 0.641]
[-3.745, -0.635]
(0.641, 3.26]
...
(-0.635, -0.022]
(0.641, 3.26]
(-0.635, -0.022]
Levels (4): Index(['[-3.745, -0.635]', '(-0.635, -0.022]',
'(-0.022, 0.641]', '(0.641, 3.26]'], dtype=object)
Length: 1000
In [168]: pd.value_counts(cats)
Out[168]:
(0.641, 3.26] 250
[-3.745, -0.635] 250
(-0.635, -0.022] 250
(-0.022, 0.641] 250
dtype: int64Similar to cut you can pass your
own quantiles (numbers between 0 and 1, inclusive):
In [169]: pd.qcut(data, [0, 0.1, 0.5, 0.9, 1.])
Out[169]:
(-0.022, 1.302]
(-1.266, -0.022]
(-0.022, 1.302]
...
(-1.266, -0.022]
(-0.022, 1.302]
(-1.266, -0.022]
Levels (4): Index(['[-3.745, -1.266]', '(-1.266, -0.022]',
'(-0.022, 1.302]', '(1.302, 3.26]'], dtype=object)
Length: 1000We’ll return to cut and qcut later in the
chapter on aggregation and group operations, as these discretization
functions are especially useful for quantile and group
analysis.
Filtering or transforming outliers is largely a matter of applying array operations. Consider a DataFrame with some normally distributed data:
In [170]: np.random.seed(12345)
In [171]: data = DataFrame(np.random.randn(1000, 4))
In [172]: data.describe()
Out[172]:
0 1 2 3
count 1000.000000 1000.000000 1000.000000 1000.000000
mean -0.067684 0.067924 0.025598 -0.002298
std 0.998035 0.992106 1.006835 0.996794
min -3.428254 -3.548824 -3.184377 -3.745356
25% -0.774890 -0.591841 -0.641675 -0.644144
50% -0.116401 0.101143 0.002073 -0.013611
75% 0.616366 0.780282 0.680391 0.654328
max 3.366626 2.653656 3.260383 3.927528Suppose you wanted to find values in one of the columns exceeding three in magnitude:
In [173]: col = data[3] In [174]: col[np.abs(col) > 3] Out[174]: 97 3.927528 305 -3.399312 400 -3.745356 Name: 3, dtype: float64
To select all rows having a value exceeding 3 or -3, you can use
the any method on a boolean
DataFrame:
In [175]: data[(np.abs(data) > 3).any(1)]
Out[175]:
0 1 2 3
5 -0.539741 0.476985 3.248944 -1.021228
97 -0.774363 0.552936 0.106061 3.927528
102 -0.655054 -0.565230 3.176873 0.959533
305 -2.315555 0.457246 -0.025907 -3.399312
324 0.050188 1.951312 3.260383 0.963301
.. ... ... ... ...
499 -0.293333 -0.242459 -3.056990 1.918403
523 -3.428254 -0.296336 -0.439938 -0.867165
586 0.275144 1.179227 -3.184377 1.369891
808 -0.362528 -3.548824 1.553205 -2.186301
900 3.366626 -2.372214 0.851010 1.332846
[11 rows x 4 columns]Values can just as easily be set based on these criteria. Here is code to cap values outside the interval -3 to 3:
In [176]: data[np.abs(data) > 3] = np.sign(data) * 3
In [177]: data.describe()
Out[177]:
0 1 2 3
count 1000.000000 1000.000000 1000.000000 1000.000000
mean -0.067623 0.068473 0.025153 -0.002081
std 0.995485 0.990253 1.003977 0.989736
min -3.000000 -3.000000 -3.000000 -3.000000
25% -0.774890 -0.591841 -0.641675 -0.644144
50% -0.116401 0.101143 0.002073 -0.013611
75% 0.616366 0.780282 0.680391 0.654328
max 3.000000 2.653656 3.000000 3.000000The ufunc np.sign returns an
array of 1 and -1 depending on the sign of the values.
Permuting (randomly reordering) a Series or the rows in a
DataFrame is easy to do using the numpy.random.permutation function. Calling
permutation with the length of the
axis you want to permute produces an array of integers indicating the
new ordering:
In [178]: df = DataFrame(np.arange(5 * 4).reshape((5, 4))) In [179]: sampler = np.random.permutation(5) In [180]: sampler Out[180]: array([1, 0, 2, 3, 4])
That array can then be used in ix-based indexing or the take function:
In [181]: df In [182]: df.take(sampler)
Out[181]: Out[182]:
0 1 2 3 0 1 2 3
0 0 1 2 3 1 4 5 6 7
1 4 5 6 7 0 0 1 2 3
2 8 9 10 11 2 8 9 10 11
3 12 13 14 15 3 12 13 14 15
4 16 17 18 19 4 16 17 18 19To select a random subset without replacement, one way is to slice
off the first k elements of the array
returned by permutation, where
k is the desired subset size. There
are much more efficient sampling-without-replacement algorithms, but
this is an easy strategy that uses readily available tools:
In [183]: df.take(np.random.permutation(len(df))[:3])
Out[183]:
0 1 2 3
1 4 5 6 7
3 12 13 14 15
4 16 17 18 19To generate a sample with replacement, the
fastest way is to use np.random.randint to
draw random integers:
In [184]: bag = np.array([5, 7, -1, 6, 4]) In [185]: sampler = np.random.randint(0, len(bag), size=10) In [186]: sampler Out[186]: array([4, 4, 2, 2, 2, 0, 3, 0, 4, 1]) In [187]: draws = bag.take(sampler) In [188]: draws Out[188]: array([ 4, 4, -1, -1, -1, 5, 6, 5, 4, 7])
Another type of transformation for statistical modeling or
machine learning applications is converting a categorical variable into
a “dummy” or “indicator” matrix. If a column in a DataFrame has k distinct values, you would derive a matrix
or DataFrame containing k columns
containing all 1’s and 0’s. pandas has a get_dummies function
for doing this, though devising one yourself is not difficult. Let’s
return to an earlier example DataFrame:
In [189]: df = DataFrame({'key': ['b', 'b', 'a', 'c', 'a', 'b'],
.....: 'data1': range(6)})
In [190]: pd.get_dummies(df['key'])
Out[190]:
a b c
0 0 1 0
1 0 1 0
2 1 0 0
3 0 0 1
4 1 0 0
5 0 1 0In some cases, you may want to add a prefix to the columns in the
indicator DataFrame, which can then be merged with the other data.
get_dummies has a
prefix argument for doing just this:
In [191]: dummies = pd.get_dummies(df['key'], prefix='key') In [192]: df_with_dummy = df[['data1']].join(dummies) In [193]: df_with_dummy Out[193]: data1 key_a key_b key_c 0 0 0 1 0 1 1 0 1 0 2 2 1 0 0 3 3 0 0 1 4 4 1 0 0 5 5 0 1 0
If a row in a DataFrame belongs to multiple categories, things are a bit more complicated. Let’s return to the MovieLens 1M dataset from earlier in the book:
In [194]: mnames = ['movie_id', 'title', 'genres']
In [195]: movies = pd.read_table('ch07/movies.dat', sep='::', header=None,
.....: names=mnames)
In [196]: movies[:10]
Out[196]:
movie_id title genres
0 1 Toy Story (1995) Animation|Children's|Comedy
1 2 Jumanji (1995) Adventure|Children's|Fantasy
2 3 Grumpier Old Men (1995) Comedy|Romance
3 4 Waiting to Exhale (1995) Comedy|Drama
4 5 Father of the Bride Part II (1995) Comedy
5 6 Heat (1995) Action|Crime|Thriller
6 7 Sabrina (1995) Comedy|Romance
7 8 Tom and Huck (1995) Adventure|Children's
8 9 Sudden Death (1995) Action
9 10 GoldenEye (1995) Action|Adventure|ThrillerAdding indicator variables for each genre requires a little bit of
wrangling. First, we extract the list of unique genres in the dataset
(using a nice set.union
trick):
In [197]: genre_iter = (set(x.split('|')) for x in movies.genres)
In [198]: genres = sorted(set.union(*genre_iter))Now, one way to construct the indicator DataFrame is to start with a DataFrame of all zeros:
In [199]: dummies = DataFrame(np.zeros((len(movies), len(genres))), columns=genres)
Now, iterate through each movie and set entries in each row of
dummies to 1:
In [200]: for i, gen in enumerate(movies.genres):
.....: dummies.ix[i, gen.split('|')] = 1Then, as above, you can combine this with movies:
In [201]: movies_windic = movies.join(dummies.add_prefix('Genre_'))
In [202]: movies_windic.ix[0]
Out[202]:
movie_id 1
title Toy Story (1995)
genres Animation|Children's|Comedy
...
Genre_Thriller 0
Genre_War 0
Genre_Western 0
Name: 0, Length: 21, dtype: objectNote
For much larger data, this method of constructing indicator variables with multiple membership is not especially speedy. A lower-level function leveraging the internals of the DataFrame could certainly be written.
A useful recipe for statistical applications is to combine
get_dummies with a
discretization function like cut:
In [204]: values = np.random.rand(10)
In [205]: values
Out[205]:
array([ 0.9296, 0.3164, 0.1839, 0.2046, 0.5677, 0.5955, 0.9645,
0.6532, 0.7489, 0.6536])
In [206]: bins = [0, 0.2, 0.4, 0.6, 0.8, 1]
In [207]: pd.get_dummies(pd.cut(values, bins))
Out[207]:
(0, 0.2] (0.2, 0.4] (0.4, 0.6] (0.6, 0.8] (0.8, 1]
0 0 0 0 0 1
1 0 1 0 0 0
2 1 0 0 0 0
3 0 1 0 0 0
4 0 0 1 0 0
5 0 0 1 0 0
6 0 0 0 0 1
7 0 0 0 1 0
8 0 0 0 1 0
9 0 0 0 1 0Python has long been a popular data munging language in part due to its ease-of-use for string and text processing. Most text operations are made simple with the string object’s built-in methods. For more complex pattern matching and text manipulations, regular expressions may be needed. pandas adds to the mix by enabling you to apply string and regular expressions concisely on whole arrays of data, additionally handling the annoyance of missing data.
In many string munging and scripting applications,
built-in string methods are sufficient. As an example, a comma-separated
string can be broken into pieces with split:
In [208]: val = 'a,b, guido'
In [209]: val.split(',')
Out[209]: ['a', 'b', ' guido']split is often combined
with strip to trim whitespace
(including newlines):
In [210]: pieces = [x.strip() for x in val.split(',')]
In [211]: pieces
Out[211]: ['a', 'b', 'guido']These substrings could be concatenated together with a two-colon delimiter using addition:
In [212]: first, second, third = pieces In [213]: first + '::' + second + '::' + third Out[213]: 'a::b::guido
But, this isn’t a practical generic method. A faster and more
Pythonic way is to pass a list or tuple to the join method on the
string '::':
In [214]: '::'.join(pieces) Out[214]: 'a::b::guido'
Other methods are concerned with locating substrings. Using
Python’s in keyword is the best way
to detect a substring, though index and find can also be
used:
In [215]: 'guido' in val
Out[215]: True
In [216]: val.index(',') In [217]: val.find(':')
Out[216]: 1 Out[217]: -1Note the difference between find and index is that
index raises an
exception if the string isn’t found (versus returning -1):
In [218]: val.index(':')
---------------------------------------------------------------------------
ValueError Traceback (most recent call last)
<ipython-input-218-280f8b2856ce> in <module>()
----> 1 val.index(':')
ValueError: substring not foundRelatedly, count returns the
number of occurrences of a particular substring:
In [219]: val.count(',')
Out[219]: 2replace will substitute
occurrences of one pattern for another. This is commonly used to delete
patterns, too, by passing an empty string:
In [220]: val.replace(',', '::') In [221]: val.replace(',', '')
Out[220]: 'a::b:: guido' Out[221]: 'ab guido'Regular expressions can also be used with many of these operations as you’ll see below.
Table 7-3. Python built-in string methods
Regular expressions provide a
flexible way to search or match string patterns in text. A single
expression, commonly called a regex, is a string formed according
to the regular expression language. Python’s built-in re module is
responsible for applying regular expressions to strings; I’ll give a
number of examples of its use here.
Note
The art of writing regular expressions could be a chapter of its own and thus is outside the book’s scope. There are many excellent tutorials and references on the internet, such as Zed Shaw’s Learn Regex The Hard Way (http://regex.learncodethehardway.org/book/).
The re module functions
fall into three categories: pattern matching, substitution, and
splitting. Naturally these are all related; a regex describes a pattern
to locate in the text, which can then be used for many purposes. Let’s
look at a simple example: suppose I wanted to split a string with a
variable number of whitespace characters (tabs, spaces, and newlines).
The regex describing one or more whitespace characters is s+:
In [222]: import re
In [223]: text = "foo bar baz qux"
In [224]: re.split('s+', text)
Out[224]: ['foo', 'bar', 'baz', 'qux']When you call re.split('s+',
text), the regular expression is first
compiled, then its split method is called on the passed text. You
can compile the regex yourself with re.compile, forming a
reusable regex object:
In [225]: regex = re.compile('s+')
In [226]: regex.split(text)
Out[226]: ['foo', 'bar', 'baz', 'qux']If, instead, you wanted to get a list of all patterns matching the
regex, you can use the findall method:
In [227]: regex.findall(text) Out[227]: [' ', ' ', ' ']
Note
To avoid unwanted escaping with in a regular expression, use
raw string literals like r'C:x' instead of the equivalent 'C:\x'.
Creating a regex object with re.compile is highly
recommended if you intend to apply the same expression to many strings;
doing so will save CPU cycles.
match and search are closely
related to findall. While findall returns all matches in a string,
search returns only the
first match. More rigidly, match
only matches at the beginning of the string. As a
less trivial example, let’s consider a block of text and a regular
expression capable of identifying most email addresses:
text = """Dave [email protected] Steve [email protected] Rob [email protected] Ryan [email protected] """ pattern = r'[A-Z0-9._%+-]+@[A-Z0-9.-]+.[A-Z]{2,4}' # re.IGNORECASE makes the regex case-insensitive regex = re.compile(pattern, flags=re.IGNORECASE)
Using findall on the text
produces a list of the e-mail addresses:
In [229]: regex.findall(text) Out[229]: ['[email protected]', '[email protected]', '[email protected]', '[email protected]']
search returns a
special match object for the first email address in the text. For the
above regex, the match object can only tell us the start and end
position of the pattern in the string:
In [230]: m = regex.search(text) In [231]: m Out[231]: <_sre.SRE_Match at 0x7fb006dfbf38> In [232]: text[m.start():m.end()] Out[232]: '[email protected]'
regex.match returns None, as it only will match if the pattern
occurs at the start of the string:
In [233]: print regex.match(text) None
Relatedly, sub will return a new
string with occurrences of the pattern replaced by the a new
string:
In [234]: print regex.sub('REDACTED', text)
Dave REDACTED
Steve REDACTED
Rob REDACTED
Ryan REDACTEDSuppose you wanted to find email addresses and simultaneously segment each address into its 3 components: username, domain name, and domain suffix. To do this, put parentheses around the parts of the pattern to segment:
In [235]: pattern = r'([A-Z0-9._%+-]+)@([A-Z0-9.-]+).([A-Z]{2,4})'
In [236]: regex = re.compile(pattern, flags=re.IGNORECASE)A match object produced by this modified regex returns a tuple of
the pattern components with its groups method:
In [237]: m = regex.match('[email protected]')
In [238]: m.groups()
Out[238]: ('wesm', 'bright', 'net')findall returns a list of
tuples when the pattern has groups:
In [239]: regex.findall(text)
Out[239]:
[('dave', 'google', 'com'),
('steve', 'gmail', 'com'),
('rob', 'gmail', 'com'),
('ryan', 'yahoo', 'com')]sub also has access to
groups in each match using special symbols like 1, 2, etc.:
In [240]: print regex.sub(r'Username: 1, Domain: 2, Suffix: 3', text) Dave Username: dave, Domain: google, Suffix: com Steve Username: steve, Domain: gmail, Suffix: com Rob Username: rob, Domain: gmail, Suffix: com Ryan Username: ryan, Domain: yahoo, Suffix: com
There is much more to regular expressions in Python, most of which is outside the book’s scope. To give you a flavor, one variation on the above email regex gives names to the match groups:
regex = re.compile(r"""
(?P<username>[A-Z0-9._%+-]+)
@
(?P<domain>[A-Z0-9.-]+)
.
(?P<suffix>[A-Z]{2,4})""", flags=re.IGNORECASE|re.VERBOSE)The match object produced by such a regex can produce a handy dict with the specified group names:
In [242]: m = regex.match('[email protected]')
In [243]: m.groupdict()
Out[243]: {'domain': 'bright', 'suffix': 'net', 'username': 'wesm'}Table 7-4. Regular expression methods
Cleaning up a messy data set for analysis often requires a lot of string munging and regularization. To complicate matters, a column containing strings will sometimes have missing data:
In [244]: data = {'Dave': '[email protected]', 'Steve': '[email protected]',
.....: 'Rob': '[email protected]', 'Wes': np.nan}
In [245]: data = Series(data)
In [246]: data In [247]: data.isnull()
Out[246]: Out[247]:
Dave [email protected] Dave False
Rob [email protected] Rob False
Steve [email protected] Steve False
Wes NaN Wes True
dtype: object dtype: boolString and regular expression methods can be applied (passing a
lambda or other
function) to each value using data.map, but it will
fail on the NA. To cope with this, Series has concise methods for string
operations that skip NA values. These are accessed through Series’s
str attribute; for example, we could
check whether each email address has 'gmail' in it with str.contains:
In [248]: data.str.contains('gmail')
Out[248]:
Dave False
Rob True
Steve True
Wes NaN
dtype: objectRegular expressions can be used, too, along with any re options like IGNORECASE:
In [249]: pattern
Out[249]: '([A-Z0-9._%+-]+)@([A-Z0-9.-]+)\.([A-Z]{2,4})'
In [250]: data.str.findall(pattern, flags=re.IGNORECASE)
Out[250]:
Dave [(dave, google, com)]
Rob [(rob, gmail, com)]
Steve [(steve, gmail, com)]
Wes NaN
dtype: objectThere are a couple of ways to do vectorized element retrieval.
Either use str.get or index into the
str attribute:
In [251]: matches = data.str.match(pattern, flags=re.IGNORECASE) In [252]: matches Out[252]: Dave (dave, google, com) Rob (rob, gmail, com) Steve (steve, gmail, com) Wes NaN dtype: object In [253]: matches.str.get(1) In [254]: matches.str[0] Out[253]: Out[254]: Dave google Dave dave Rob gmail Rob rob Steve gmail Steve steve Wes NaN Wes NaN dtype: object dtype: object
You can similarly slice strings using this syntax:
In [255]: data.str[:5] Out[255]: Dave dave@ Rob rob@g Steve steve Wes NaN dtype: object
Table 7-5. Vectorized string methods
The US Department of Agriculture makes available a database of food nutrient information. Ashley Williams, an English hacker, has made available a version of this database in JSON format (http://ashleyw.co.uk/project/food-nutrient-database). The records look like this:
{
"id": 21441,
"description": "KENTUCKY FRIED CHICKEN, Fried Chicken, EXTRA CRISPY,
Wing, meat and skin with breading",
"tags": ["KFC"],
"manufacturer": "Kentucky Fried Chicken",
"group": "Fast Foods",
"portions": [
{
"amount": 1,
"unit": "wing, with skin",
"grams": 68.0
},
...
],
"nutrients": [
{
"value": 20.8,
"units": "g",
"description": "Protein",
"group": "Composition"
},
...
]
}
Each food has a number of identifying attributes along with two lists of nutrients and portion sizes. Having the data in this form is not particularly amenable for analysis, so we need to do some work to wrangle the data into a better form.
After downloading and extracting the data from the link above, you
can load it into Python with any JSON library of your choosing. I’ll use
the built-in Python json module:
In [256]: import json
In [257]: db = json.load(open('ch07/foods-2011-10-03.json'))
In [258]: len(db)
Out[258]: 6636Each entry in db is a dict
containing all the data for a single food. The 'nutrients' field is a list of dicts, one for
each nutrient:
In [259]: db[0].keys() In [260]: db[0]['nutrients'][0]
Out[259]: Out[260]:
[u'portions', {u'description': u'Protein',
u'description', u'group': u'Composition',
u'tags', u'units': u'g',
u'nutrients', u'value': 25.18}
u'group',
u'id',
u'manufacturer']
In [261]: nutrients = DataFrame(db[0]['nutrients'])
In [262]: nutrients[:7]
Out[262]:
description group units value
0 Protein Composition g 25.18
1 Total lipid (fat) Composition g 29.20
2 Carbohydrate, by difference Composition g 3.06
3 Ash Other g 3.28
4 Energy Energy kcal 376.00
5 Water Composition g 39.28
6 Energy Energy kJ 1573.00When converting a list of dicts to a DataFrame, we can specify a list of fields to extract. We’ll take the food names, group, id, and manufacturer:
In [263]: info_keys = ['description', 'group', 'id', 'manufacturer']
In [264]: info = DataFrame(db, columns=info_keys)
In [265]: info[:5]
Out[265]:
description group id
0 Cheese, caraway Dairy and Egg Products 1008
1 Cheese, cheddar Dairy and Egg Products 1009
2 Cheese, edam Dairy and Egg Products 1018
3 Cheese, feta Dairy and Egg Products 1019
4 Cheese, mozzarella, part skim milk Dairy and Egg Products 1028
manufacturer
0
1
2
3
4
In [266]: info.info()
<class 'pandas.core.frame.DataFrame'>
Int64Index: 6636 entries, 0 to 6635
Data columns (total 4 columns):
description 6636 non-null object
group 6636 non-null object
id 6636 non-null int64
manufacturer 5195 non-null object
dtypes: int64(1), object(3)You can see the distribution of food groups with value_counts:
In [267]: pd.value_counts(info.group)[:10] Out[267]: Vegetables and Vegetable Products 812 Beef Products 618 Baked Products 496 Breakfast Cereals 403 Legumes and Legume Products 365 Fast Foods 365 Lamb, Veal, and Game Products 345 Sweets 341 Fruits and Fruit Juices 328 Pork Products 328 dtype: int64
Now, to do some analysis on all of the nutrient data, it’s easiest
to assemble the nutrients for each food into a single large table. To do
so, we need to take several steps. First, I’ll convert each list of food
nutrients to a DataFrame, add a column for the food id, and append the DataFrame to a list. Then,
these can be concatenated together with concat:
nutrients = []
for rec in db:
fnuts = DataFrame(rec['nutrients'])
fnuts['id'] = rec['id']
nutrients.append(fnuts)
nutrients = pd.concat(nutrients, ignore_index=True)If all goes well, nutrients
should look like this:
In [269]: nutrients
Out[269]:
description group units value id
0 Protein Composition g 25.180 1008
1 Total lipid (fat) Composition g 29.200 1008
2 Carbohydrate, by difference Composition g 3.060 1008
3 Ash Other g 3.280 1008
4 Energy Energy kcal 376.000 1008
... ... ... ... ... ...
389350 Vitamin B-12, added Vitamins mcg 0.000 43546
389351 Cholesterol Other mg 0.000 43546
389352 Fatty acids, total saturated Other g 0.072 43546
389353 Fatty acids, total monounsaturated Other g 0.028 43546
389354 Fatty acids, total polyunsaturated Other g 0.041 43546
[389355 rows x 5 columns]I noticed that, for whatever reason, there are duplicates in this DataFrame, so it makes things easier to drop them:
In [270]: nutrients.duplicated().sum() Out[270]: 14179 In [271]: nutrients = nutrients.drop_duplicates()
Since 'group' and 'description' is in both DataFrame objects, we
can rename them to make it clear what is what:
In [272]: col_mapping = {'description' : 'food',
.....: 'group' : 'fgroup'}
In [273]: info = info.rename(columns=col_mapping, copy=False)
In [274]: info.info()
<class 'pandas.core.frame.DataFrame'>
Int64Index: 6636 entries, 0 to 6635
Data columns (total 4 columns):
food 6636 non-null object
fgroup 6636 non-null object
id 6636 non-null int64
manufacturer 5195 non-null object
dtypes: int64(1), object(3)
In [275]: col_mapping = {'description' : 'nutrient',
.....: 'group' : 'nutgroup'}
In [276]: nutrients = nutrients.rename(columns=col_mapping, copy=False)
In [277]: nutrients
Out[277]:
nutrient nutgroup units value id
0 Protein Composition g 25.180 1008
1 Total lipid (fat) Composition g 29.200 1008
2 Carbohydrate, by difference Composition g 3.060 1008
3 Ash Other g 3.280 1008
4 Energy Energy kcal 376.000 1008
... ... ... ... ... ...
389350 Vitamin B-12, added Vitamins mcg 0.000 43546
389351 Cholesterol Other mg 0.000 43546
389352 Fatty acids, total saturated Other g 0.072 43546
389353 Fatty acids, total monounsaturated Other g 0.028 43546
389354 Fatty acids, total polyunsaturated Other g 0.041 43546
[375176 rows x 5 columns]With all of this done, we’re ready to merge info with nutrients:
In [278]: ndata = pd.merge(nutrients, info, on='id', how='outer') In [279]: ndata.info() <class 'pandas.core.frame.DataFrame'> Int64Index: 375176 entries, 0 to 375175 Data columns (total 8 columns): nutrient 375176 non-null object nutgroup 375176 non-null object units 375176 non-null object value 375176 non-null float64 id 375176 non-null int64 food 375176 non-null object fgroup 375176 non-null object manufacturer 293054 non-null object dtypes: float64(1), int64(1), object(6) In [280]: ndata.ix[30000] Out[280]: nutrient Glycine nutgroup Amino Acids units g value 0.04 id 6158 food Soup, tomato bisque, canned, condensed fgroup Soups, Sauces, and Gravies manufacturer Name: 30000, dtype: object
The tools that you need to slice and dice, aggregate, and visualize this dataset will be explored in detail in the next two chapters, so after you get a handle on those methods you might return to this dataset. For example, we could a plot of median values by food group and nutrient type (see Figure 7-1):
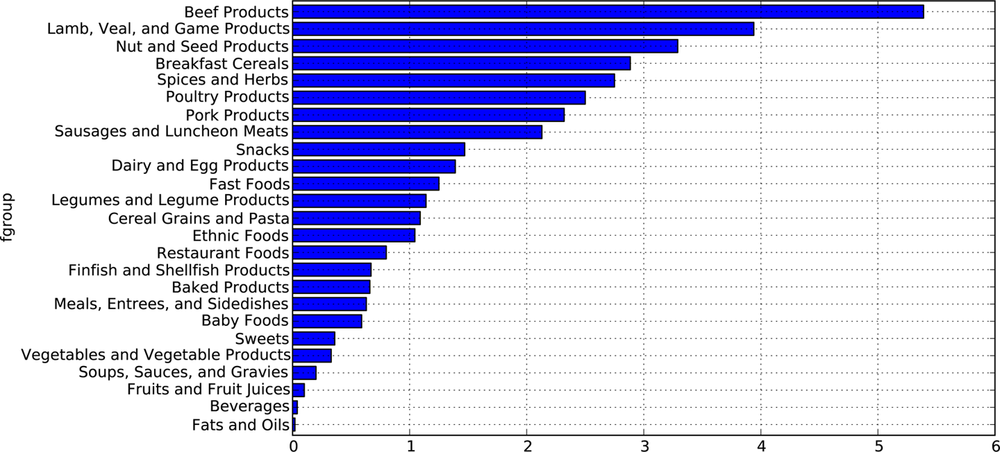
In [281]: result = ndata.groupby(['nutrient', 'fgroup'])['value'].quantile(0.5) In [282]: result['Zinc, Zn'].order().plot(kind='barh')
With a little cleverness, you can find which food is most dense in each nutrient:
by_nutrient = ndata.groupby(['nutgroup', 'nutrient']) get_maximum = lambda x: x.xs(x.value.idxmax()) get_minimum = lambda x: x.xs(x.value.idxmin()) max_foods = by_nutrient.apply(get_maximum)[['value', 'food']] # make the food a little smaller max_foods.food = max_foods.food.str[:50]
The resulting DataFrame is a bit too large to display in the book;
here is just the 'Amino Acids' nutrient
group:
In [284]: max_foods.ix['Amino Acids']['food'] Out[284]: nutrient Alanine Gelatins, dry powder, unsweetened Arginine Seeds, sesame flour, low-fat Aspartic acid Soy protein isolate ... Tryptophan Sea lion, Steller, meat with fat (Alaska Native) Tyrosine Soy protein isolate, PROTEIN TECHNOLOGIES INTE... Valine Soy protein isolate, PROTEIN TECHNOLOGIES INTE... Name: food, Length: 19, dtype: object