In this chapter, we will look at the basics of data management through the Wolfram Data Repository online platform. We will review how this website is built in order to have a better understanding of its use in Mathematica through the Wolfram Language. Examples will be carried out on how to download the data from this platform through the use of the Wolfram Language as well as its representation of data in the form dataset as well as using the Query command. We will also look at how data can be viewed inside datasets, how to apply user functions, and commands inside the format dataset.
Wolfram Data Repository
The Wolfram data repository is a website, which in turn is a repository of data, which is in the Cloud. This data repository contains information from different categories, such as computer science, meteorology, agriculture, sports, text and literature education, and many more. Although this repository belongs to Wolfram Research, it is characterized by being of public domain. In the Wolfram Data Repository, the information contained is computable data that has been selected, structured, and cured to be for direct use, to perform numerical calculations, estimates, analysis, statistics, or demonstrations, among others. The content hosted in this repository is data from many sources, globally known datasets, and publication data. All this information is designed so that any individual can access it globally. The Wolfram Data Repository system provides a data source that, in turn, also enables the storage of new information. The information that is stored in the repository is designed for direct implementation to the Wolfram Language.

Http response object of the Wolfram Data Repository. As we can see, we have received a successful response
Wolfram Data Repository Website

Wolfram Data Repository website
The images that appear are links that redirect to the dataset associated with that image.
Life Science category of the Wolfram Data Repository
The same process is for when we navigate by data type.
Selecting a Category

Fisher’s Irises dataset

Wolfram Cloud sign-in prompt
In the new window, you will enter your email and password to be able to access from Mathematica the contents of the Wolfram Data Repository.
Extracting Data from the Wolfram Data Repository
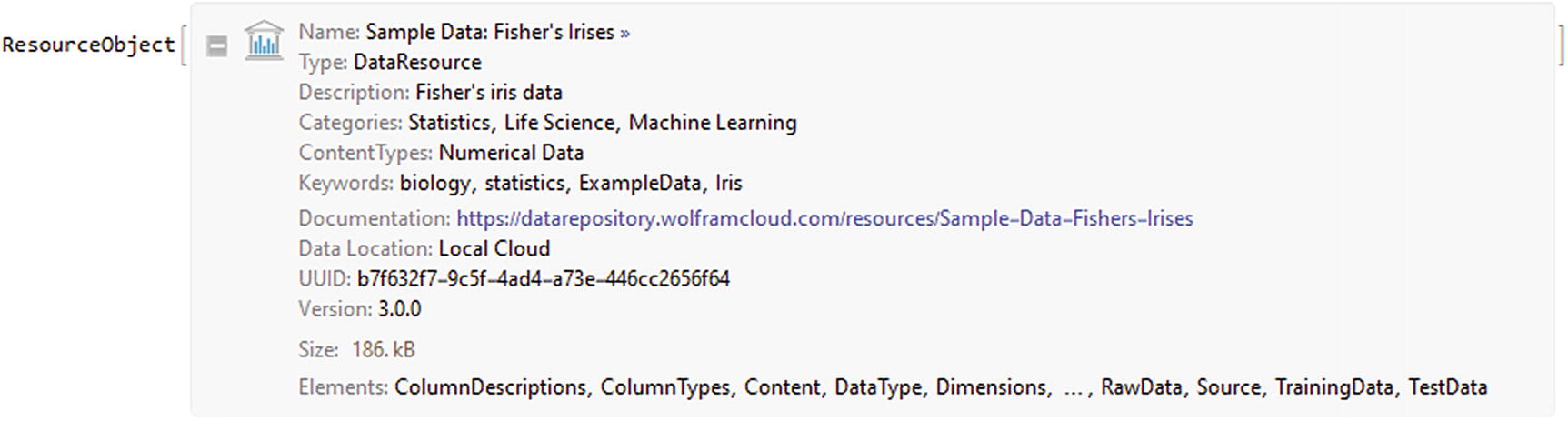
ResourceObject of the Fisher´s Irises
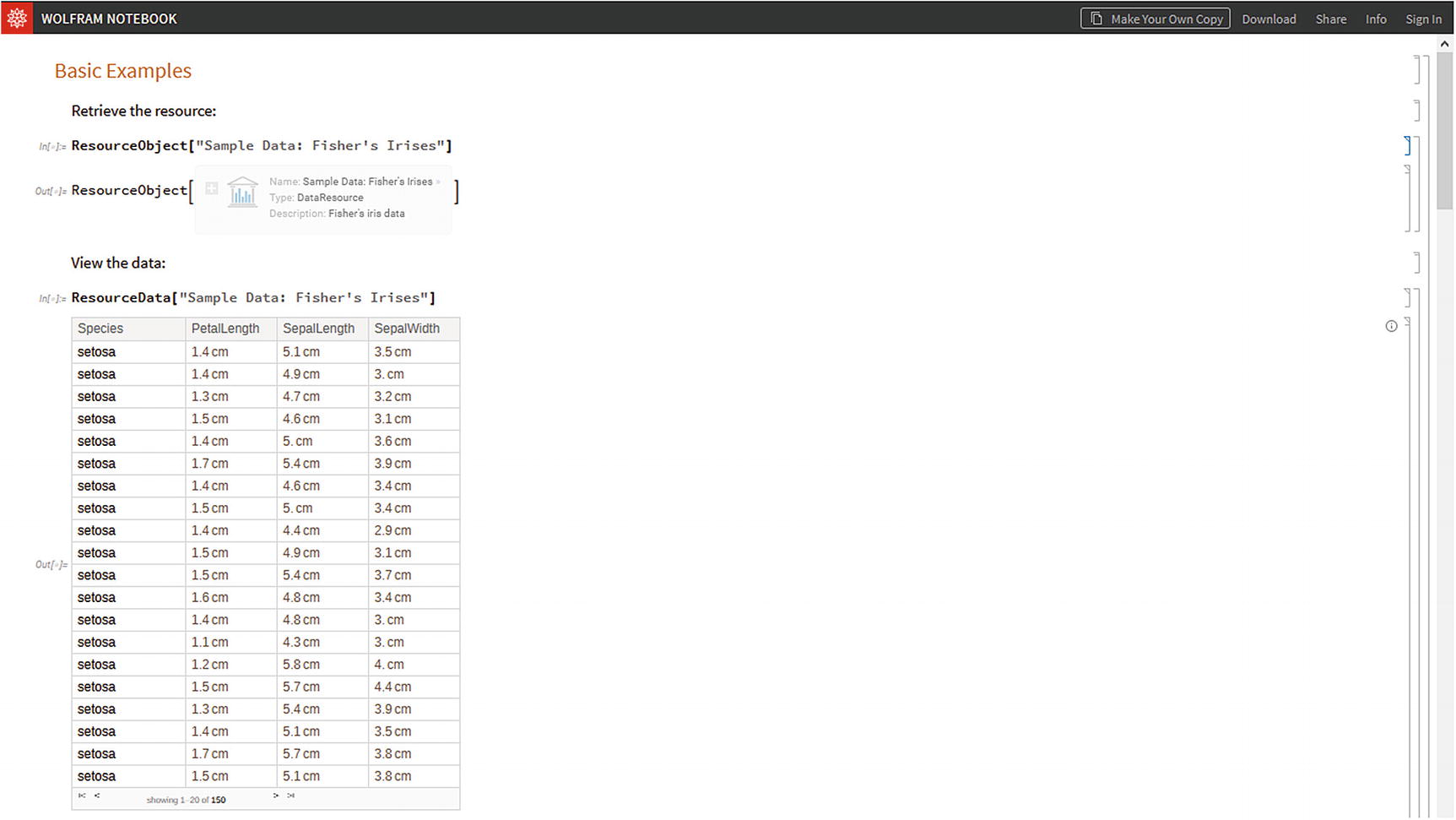
Fisher´s Irises data sample, open from the Wolfram Cloud
Accessing Data Inside Mathematica
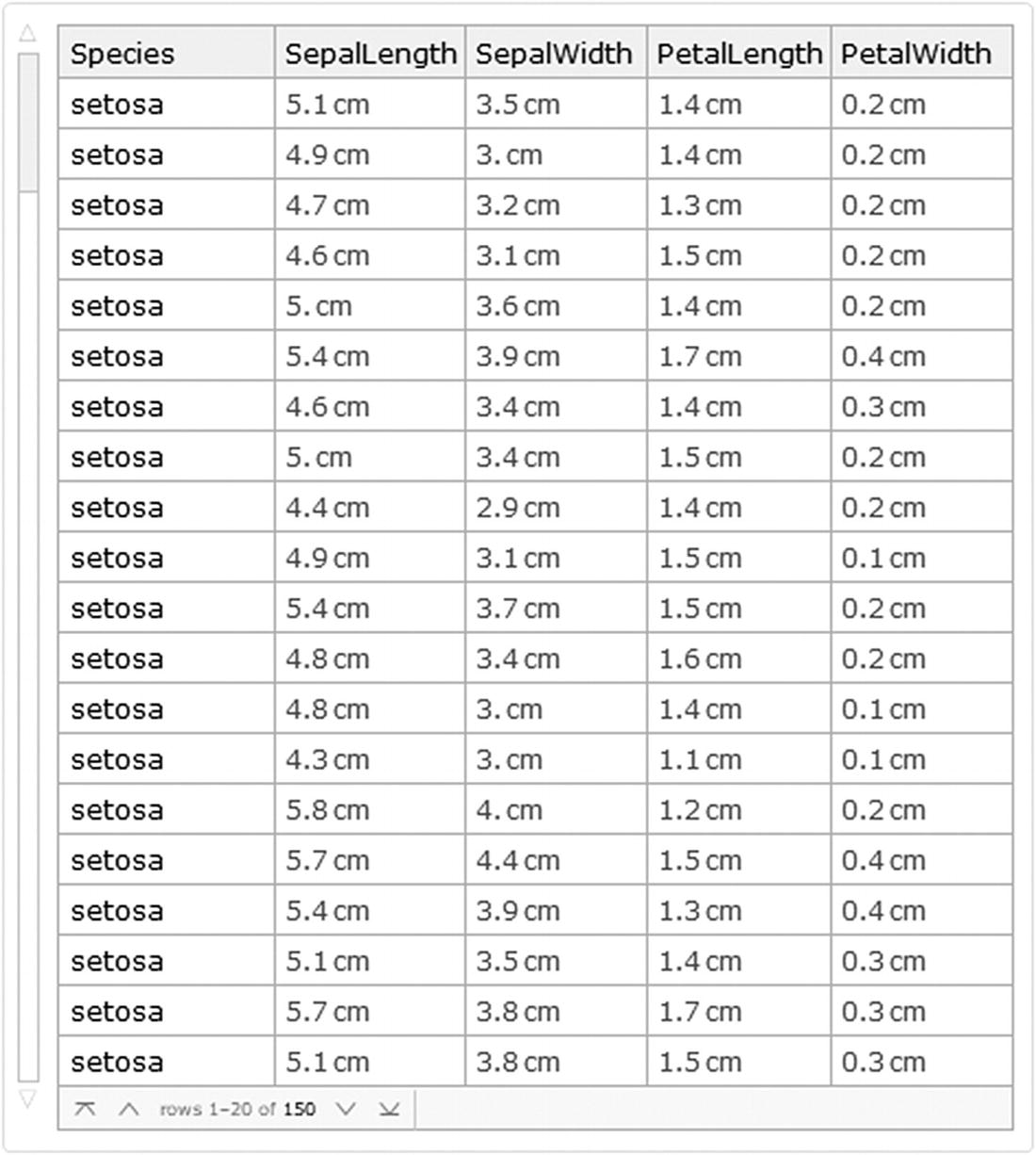
Fisher’s Irises dataset object
This way we get to know the type of information in the columns ,such as dimensions, which are 150 rows per 4 columns and the data source.
Data Observation
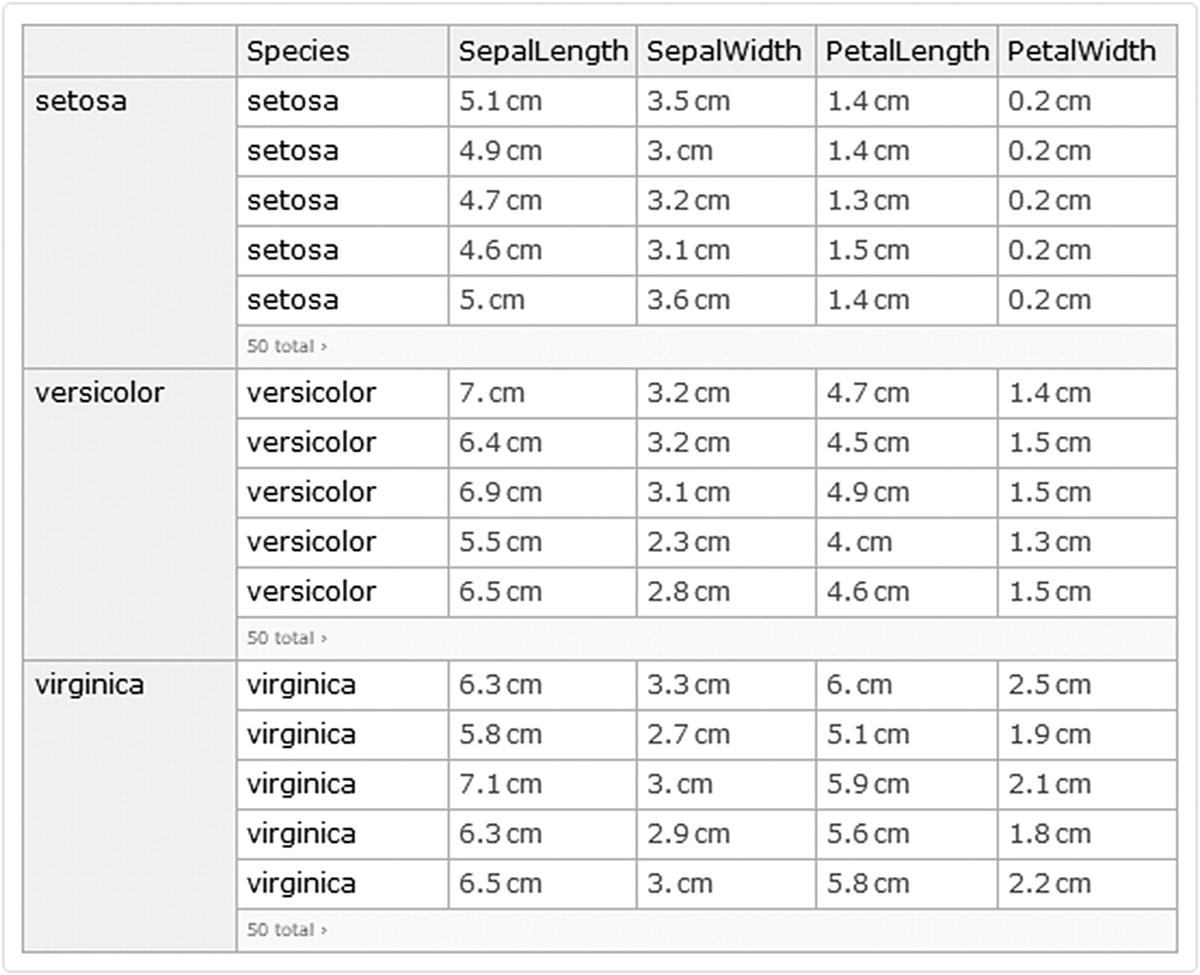
Iris data grouped by species
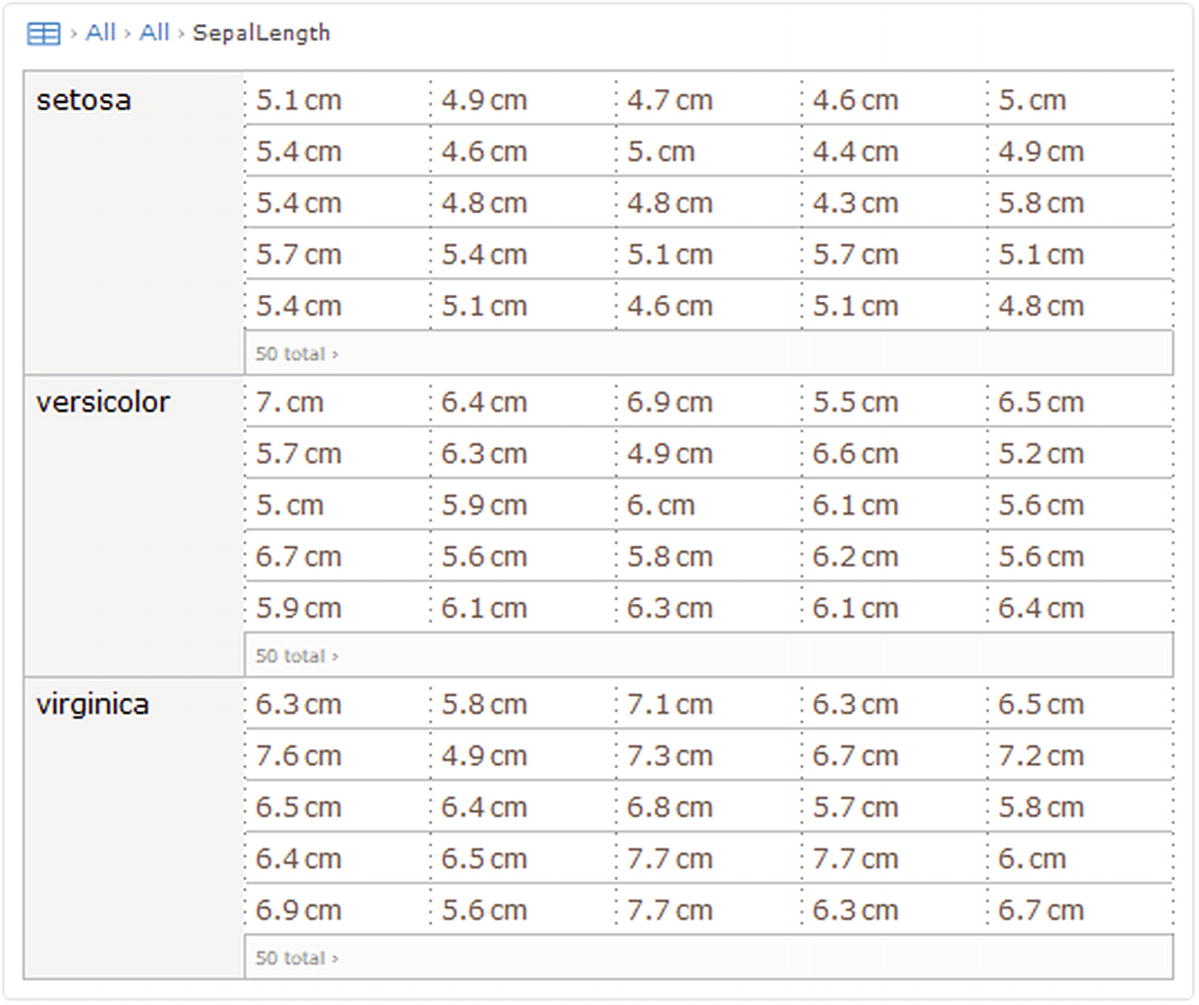
SepalLength column selected
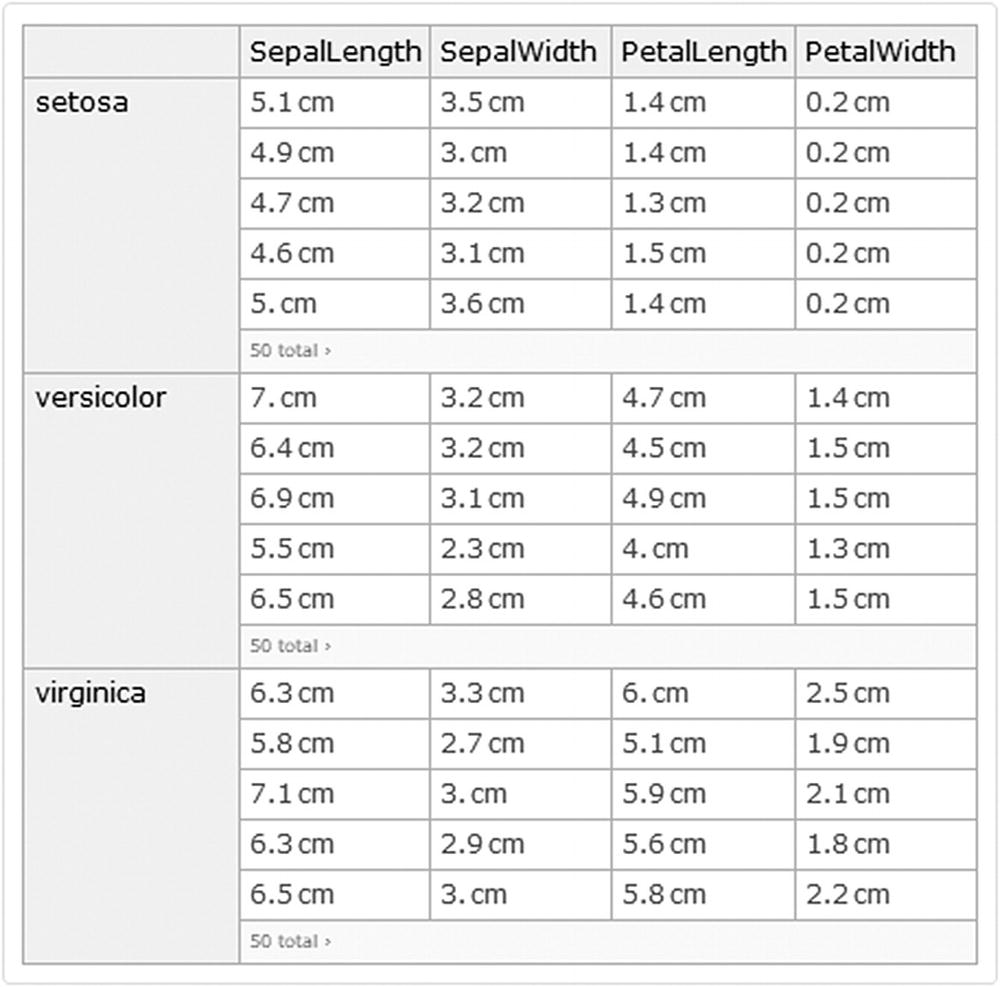
Dataset with the species column suppressed
What happens in the latter code is that we use the Key command to access the keys of the species column. Once these keys are accessed, we write a transformation rule so that each extracted key is assigned the associations extracted (KeyTake) from columns (SepalLength, SepalWidth, PetalLength, PetalWidth), then grouped and applied to Fisher’s dataset.

ID's added to the Fisher´s dataset
If we drag down the bar, we see that the counter reaches 150 elements.

Counted elements on the dataset
This results in 50 data belonging to setosa, versicolor, and virginica. If we add them up, we get 150. You can also use the Query command, Query[Counts, "Species"][Fisher].

Mean for the four columns, divided by species
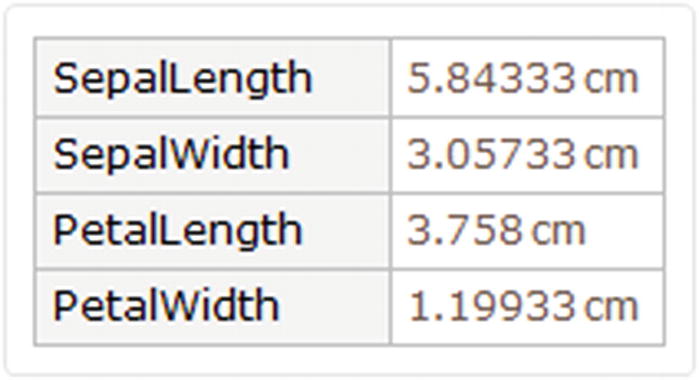
Average values for the four columns of all species
The Mean command works with the quantities and returns the average to use as a quantity.
Descriptive Statistics

Function Stats applied to each column

Tabview format
With TabView, we create three tabs with the names of each column, where it shows the values maximum, minimum, average, median, first, and third quartile; the columns are SepalLength, SepalWidth, PetalLength, and PetalWidth.
Table and Grid Formats
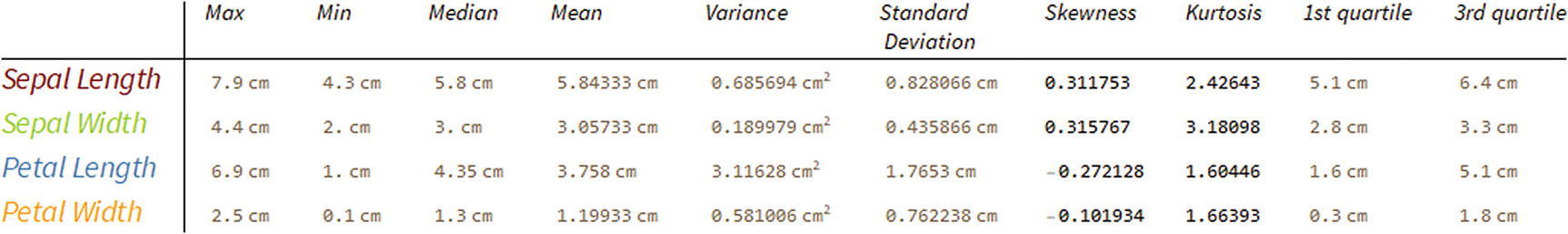
Table showing descriptive statistics by the four features

Grid view of the descriptive statistics

Descriptive stats for the versicolor specie
We have only done this for the species of versicolor; if required the same process will be performed for each species. For example, if choose Cases with the other species, we would change the text to the corresponding specie.
Dataset Visualization
Having viewed the capabilities of the Wolfram Language to perform descriptive statistics within dataset, statistical charts can be implemented inside the dataset format, as we will see in this fragment.
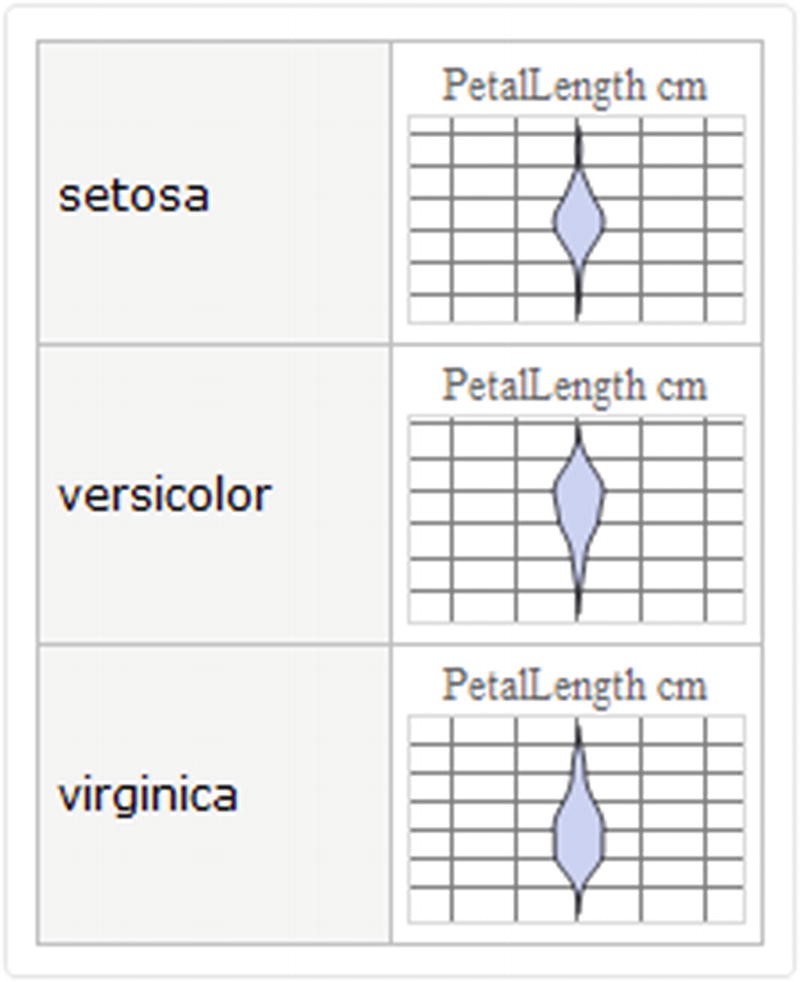
Distribution chart plot

Box whiskers plot

Box whiskers plot for virginica specie

Histogram plot for versicolor
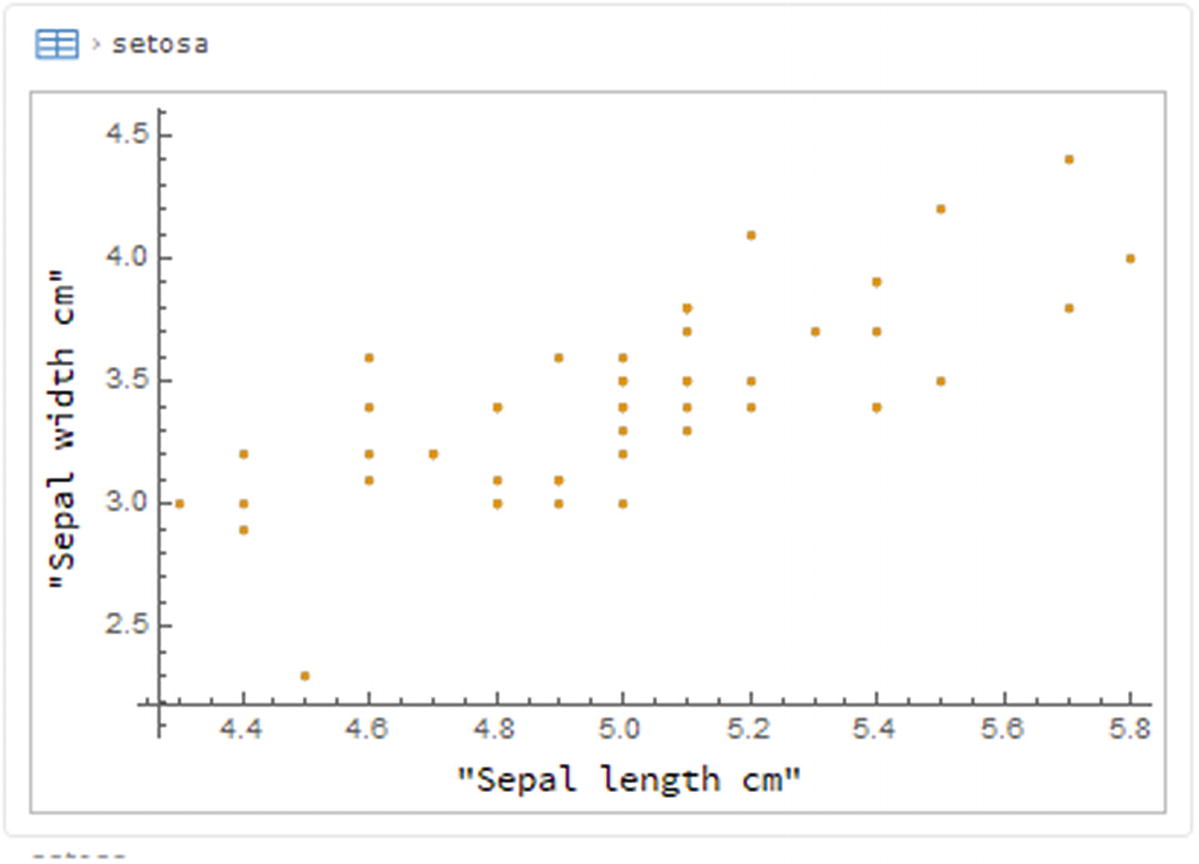
2D scatter plot
To return to the full dataset, click the dataset icon as with any other.
Data Outside Dataset Format
It's worth asking a question here: Why do we remove the units if calculations can be made with them? We extract the magnitudes for all quantities because they have the same order of magnitude (cm), so each calculation will be in the same units, except if we made conversions or transformations to the data.
2D and 3D Plots

Box whiskers plot and distribution chart for all species

Box whiskers plot for every specie with the four features

2D scatter plot for all species of the first two features
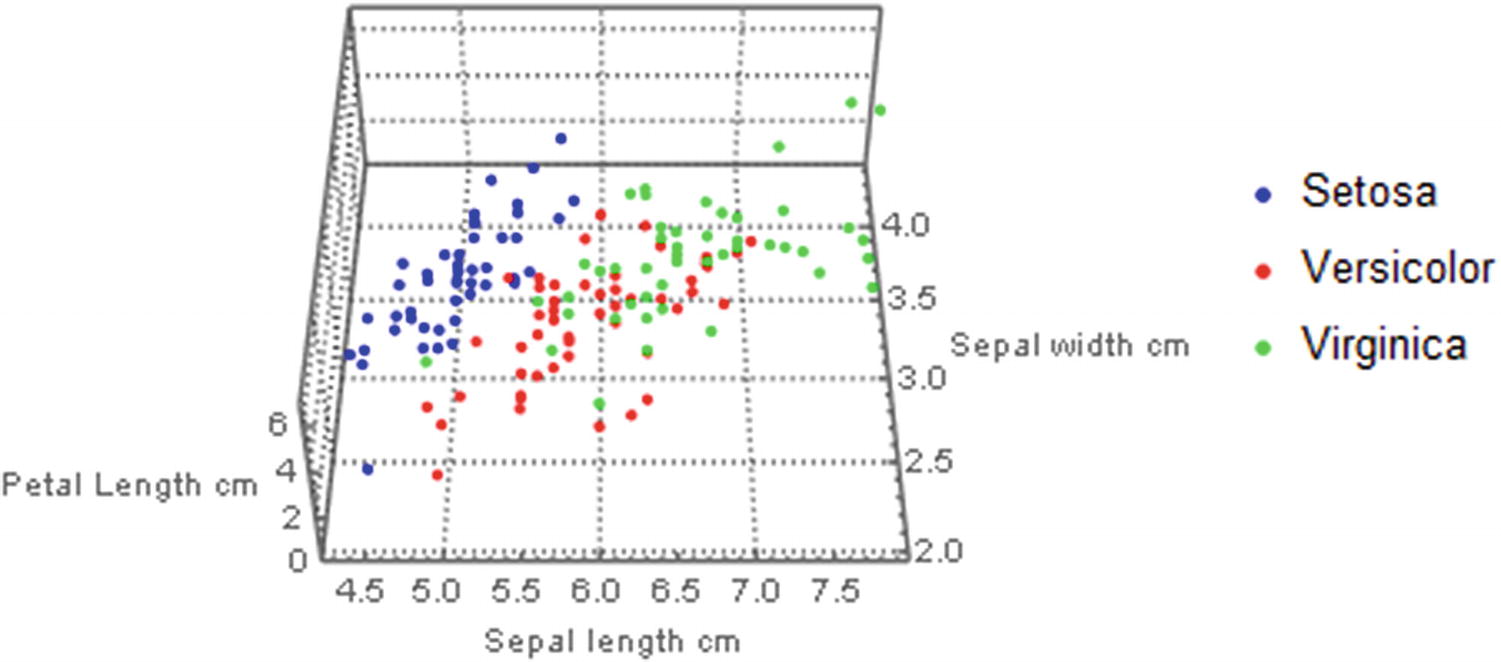
3D scatter plot of three features for every species
