As we have chosen to use GDELT for analysis purposes in this book, we will introduce our first example using this dataset. First, let's select some data.
There are two streams of data available: Global Knowledge Graph (GKG) and Events.
For this chapter, we are going to use GKG data to create a time-series dataset queryable from Spark SQL. This will give us a great starting point to create some simple introductory analytics.
In the next chapters, Chapter 4, Exploratory Data Analysis and Chapter 5, Spark for Geographic Analysis, we'll go into more detail but stay with GKG. Then, in Chapter 7, Building Communities, we will explore events by producing our own network graph of persons and using it in some cool analytics.
GDELT has been around for more than 20 years and, during that time, has undergone some significant revisions. For our introductory examples, to keep things simple, let's limit our range of data from 1st April 2013, when GDELT had a major file structure overhaul, introducing the GKG files. It's worth noting that the principles discussed in this chapter are applicable to all versions of GDELT data, however, the specific schemas and Uniform Resource Identifiers (URIs) prior to this date may be different to the ones described. The version we will use is GDELT v2.1, which is the latest version at the time of writing. But again, it's worth noting that this varies only slightly from GDELT 2.0.
There are two data tracks within GKG data:
- The entire knowledge graph, along with all of its fields.
- The subset of the graph, which contains a set of predefined categories.
We'll look at the first track.
We discussed how to download GDELT data in Chapter 2, Data Acquisition, so if you already have a NiFi pipeline configured to download the GKG data, just ensure that it's available in HDFS. However, if you have not completed that chapter, then we would encourage you to do this first, as it explains why you should take a structured approach to obtaining data.
While we have gone to great lengths to discourage the use of ad hoc data downloading, the scope of this chapter is of course known and therefore, if you are interested in following the examples seen here, you can skip the use of NiFi and obtain the data directly (in order to get started as quickly as possible).
If you do wish to download a sample, here's a reminder of where to find the GDELT 2.1 GKG master file list:
http://data.gdeltproject.org/gdeltv2/masterfilelist.txt
Make a note of a couple of the latest entries that match .gkg.csv.zip, copy them using your favorite HTTP tool, and upload them into HDFS. For example:
wget http://data.gdeltproject.org/gdeltv2/20150218230000.gkg.csv.zip -o log.txt unzip 20150218230000.gkg.csv.zip hdfs dfs -put 20150218230000.gkg.csv /data/gdelt/gkg/2015/02/21/
Now that you have unzipped your CSV file and loaded it into HDFS, let's get on and look at the data.
There are some important principles to understand which will certainly save time in the long run, whether in terms of computing or human effort. Like many CSVs, this file is hiding some complexity that, if not understood well at this stage, could become a real problem during our large scale analytics later. The GDELT documentation describes the data. It can be found here: http://data.gdeltproject.org/documentation/GDELT-Global_Knowledge_Graph_Codebook-V2.1.pdf.
It indicates that each CSV line is newline delimite, and structured as in Figure 1:

On the face of it, this appears to be a nice, simple model whereby we can simply query a field and use the enclosed data-exactly like the CSV files we import and export to Microsoft Excel every day. However, if we examine the fields in more detail, it becomes clear that some of the fields are actually references to external sources and others are flattened data, actually represented by other tables.
The flattened data structures in a core GKG model represent hidden complexity. For example, looking at field V2GCAM in the documentation, it outlines the idea that this is a series of comma-delimited blocks containing colon-delimited key-value pairs, the pairs representing GCAM variables, and their respective counts. Like so:
wc:125,c2.21:4,c10.1:40,v10.1:3.21111111
If we reference the GCAM specification, http://data.gdeltproject.org/documentation/GCAM-MASTER-CODEBOOK.TXT we can translate this to:

There are also other fields that work in the same way, such as V2Locations, V2Persons, V2Organizations, and so on. So, what's really going on here? What are all these nested structures and why would you choose to represent data in this way? Actually, it turns out that this is a convenient way to collapse a dimensional model so that it can be represented in single line records without any loss of data or cross-referencing. In fact, it's a frequently used technique, known as denormalization.
Traditionally, a dimensional model is a database table structure that comprises many fact and dimension tables. They are often referred to as having star or snowflake schemas due to their appearance in entity-relation diagrams. In such a model, a fact is a value that can be counted or summed and typically provides a measurement at a given point in time. As they are often based on transactions, or repeating events, the number of facts are prone to growing very large. A dimension on the other hand is a logical grouping of information whose purpose is to qualify or contextualize facts. They usually provide an entry point for interpreting facts by means of grouping or aggregation. Also, dimensions can be hierarchical and one dimension can reference another. We can see a diagram of the expanded GKG dimensional structure in Figure 2.
In our GCAM example, the facts are the entries found in the above table, and the dimension is the GCAM reference itself. While this may seem like a simple, logical abstraction, it does mean that we have an important area of concern that we should consider carefully: dimensional modeling is great for traditional databases where data can be split into tables–in this case, GKG and GCAM tables–as these types of databases, by their very nature, are optimized for that structure. For example, the operations for looking up values or aggregating facts are available natively. When using Spark, however, some of the operations that we take for granted can be very expensive. For example, if we wanted to average all of the GCAM fields for millions of entries, then we would have a very large computation to perform. We will discuss this in more detail in the following diagram:
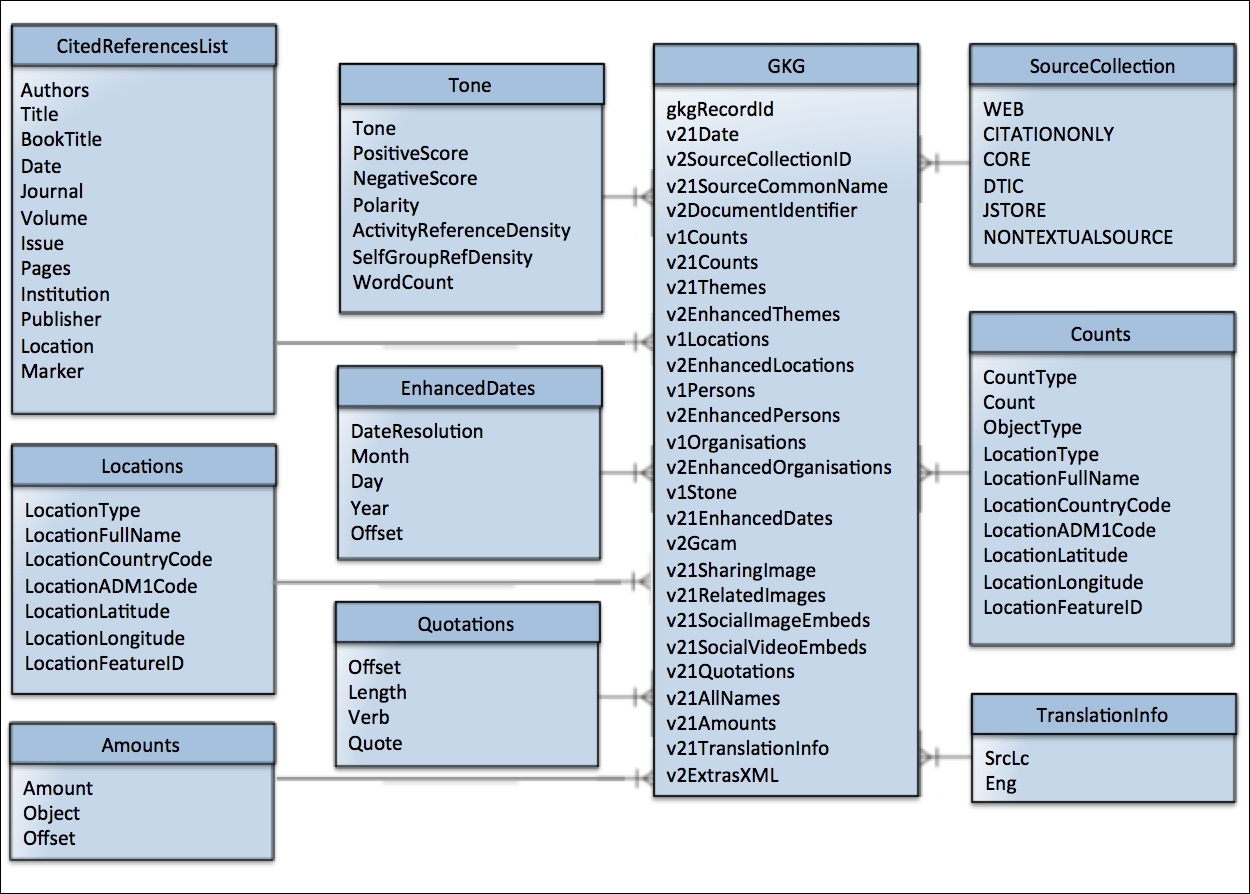
Having explored the GKG data schema, we now know that the taxonomy is a typical star schema with a single fact table referencing multiple dimension tables. With this hierarchical structure, we will certainly struggle should we need to slice-and-dice data in the same way a traditional database would allow.
But what makes it so difficult to process on Spark? Let's look at three different issues inherent with this type of organization.
First, there is the matter of the various arrays used within each record of the dataset. For example, V1Locations, V1Organizations, and V1Persons fields all contain a list of 0 or more objects. As we do not have the original body of the text used to derive this information (although we can sometimes obtain it if the source is WEB, JSTOR, and so on, since those will contain links to the source document), we lose the context of the relationships between the entities.
For example, if we have [Barack Obama, David Cameron, Francois Hollande, USA, France, GB, Texaco, Esso, Shell] in our data, then we could make the assumption that the source article is related to a meeting between heads of state over an oil crisis. However, this is only an assumption and may not be the case, if we were truly objective, we could equally assume that the article was related to companies who had employees with famous names.
To help us to infer these relationships between entities, we can develop a time series model that takes all of the individual contents of a GDELT field, over a certain time period, and performs an expansion join. Thus, on a simple level, those pairs that are seen more often are more likely to actually relate to each other and we can start to make some more concrete assumptions. For example, if we see [Barack Obama, USA] 100,000 times in our timeseries and [Barack Obama, France] only 5000 times, then it is very likely that there is a strong relationship between the first pair, and a secondary relationship between the second. In other words, we can identify the tenuous relationships and remove them when needed. This method can be used at scale to identify relationships between apparently unrelated entities. In Chapter 7, Building Communities, we use this principle to identify relationships between some very unlikely people!
With any denormalized data it should be possible to reconstruct, or inflate, the original dimensional model. With this in mind, let's look at a useful Spark function that will help us to expand our arrays and produce a flattened result; it's called DataFrame.explode, and here's an illustrative example:
case class Grouped(locations:Array[String], people:Array[String])
val group = Grouped(Array("USA","France","GB"),
Array("Barack Obama","David Cameron", "Francois Hollande"))
val ds = Seq(group).toDS
ds.show
+-----------------+--------------------+
| locations| people|
+-----------------+--------------------+
|[USA, France, GB]|[Barack Obama, Da...|
+-----------------+--------------------+
val flatLocs = ds.withColumn("locations",explode($"locations"))
flatLocs.show
+---------+--------------------+
|Locations| People|
+---------+--------------------+
| USA|[Barack Obama, Da...|
| France|[Barack Obama, Da...|
| GB|[Barack Obama, Da...|
+---------+--------------------+
val flatFolk = flatLocs.withColumn("people",explode($"people"))
flatFolk.show
+---------+-----------------+
|Locations| People|
+---------+-----------------+
| USA| Barack Obama|
| USA| David Cameron|
| USA|Francois Hollande|
| France| Barack Obama|
| France| David Cameron|
| France|Francois Hollande|
| GB| Barack Obama|
| GB| David Cameron|
| GB|Francois Hollande|
+---------+-----------------+
Using this method, we can easily expand arrays and then perform the grouping of our choice. Once expanded, the data is readily aggregated using the DataFrame methods and can even be done using SparkSQL. An example of this can be found in the Zeppelin notebooks in our repository.
It is important to understand that, while this function is simple to implement, it is not necessarily performant and may hide the underlying processing complexity required. In fact, there is an example of the explode function using GKG data within the Zeppelin notebook that accompanies this chapter, whereby, if the explode functions are not reasonably scoped, then the function returns a heap space issue as it runs out of memory.
This function does not solve the inherent problem of consuming large amounts of system resources, and so you should still take care when using it. And while this general problem cannot be solved, it can be managed by performing only the groupings and joins necessary, or by calculating them ahead of time and ensuring they complete within the resources available. You may even wish to write an algorithm that splits a dataset and performs the grouping sequentially, persisting each time. We explore methods to help us with this problem, and other common processing issues, in Chapter 14, Scalable Algorithms.
For this issue, let's look at the GDELT event data, which we have expanded in Figure 3:

This type of diagrammatic representation draws attention to the relationships in the data and gives an indication of how we might want to inflate it. Here, we see many fields that are just codes and would require translation back into their original descriptions in order to present anything meaningful. For example, in order to interpret the Actor1CountryCode (GDELT events), we will need to join the event data with one or more separate reference datasets that provide the translation text. In this case, the documentation tells us to reference the CAMEO dataset located here: http://data.gdeltproject.org/documentation/CAMEO.Manual.1.1b3.pdf.
This type of join has always presented a serious problem at data scale and there are various ways to handle it depending upon the given scenario - it is important at this stage to understand exactly how your data will be used, which joins may be required immediately, and which may be deferred until sometime in the future.
In the case where we choose to completely denormalize, or flatten, the data before processing, then it makes sense to do the join upfront. In this case, follow-on analytics will certainly be more efficient, as the relevant joins have already been completed:
So, in our example:
wc:125,c2.21:4,c10.1:40,v10.1:3.21111111
For each code in the record, there is a join to the respective reference table, and the entire record becomes:
WordCount:125, General_Inquirer_Bodypt:4, SentiWordNet:40, SentiWordNet average: v10.1:3.21111111
This is a simple change, but is one that uses a lot of disk space if performed across large numbers of rows. The trade-off is that the joins have to be performed at some point, perhaps at ingest or as a regular batch job after ingest; it is perfectly reasonable to ingest the data as is, and perform flattening of the dataset at a time that is convenient to the user. In any case, the flattened data can be consumed by any analytic and data analysts need not concern themselves with this potentially hidden issue.
On the other hand, often, deferring the join until later in the processing can mean that there are fewer records to join with – as there may have been aggregation steps in the pipeline. In this case, joining to tables at the last possible opportunity pays off because, often, the reference or dimension tables are small enough to be broadcast joins, or map-side joins. As this is such an important topic, we will continue to look at different ways of approaching join scenarios throughout the book.
