Chapter 2: Pachyderm Basics
Pachyderm is a data science platform that enables data scientists to create an end-to-end machine learning workflow that covers the most important stages of a machine learning life cycle, starting from data ingestion all the way into production.
If you are familiar with Git, a version control and life cycle system for code, you will find many similarities between the most important Git and Pachyderm concepts. Version control systems such as Git and its hosted version GitHub have become an industry standard for thousands of developers worldwide. Git enables you to keep a history of changes in your code and go back when needed. Data scientists deserve a platform that will let them track the versions of their experiments, reproduce results when needed, and investigate and correct bias that might crawl into one of the stages of the data science life cycle. Pachyderm provides benefits similar to Git that enable data scientists to reproduce their experiments and effortlessly manage the complete life cycle of the data science workflow.
This chapter is intended to describe the basics of Pachyderm architecture and its main concepts. We will cover the following topics:
- Reviewing Pachyderm architecture
- Learning about version control primitives
- Discovering pipeline elements
Reviewing Pachyderm architecture
This section walks you through the distributed Pachyderm architecture and the internals of the Pachyderm solution. But before we dive into the nitty-gritty details of Pachyderm infrastructure, let's answer the question that a lot of you might have on your mind after reading the introduction—why can't I use Git or any other version control system? We'll address this question with Git in mind as it is the most popular and widely used software version control system, but all of the arguments apply to any other similar version control system for source code. After we review how Pachyderm is different and similar to Git, we will review the Pachyderm internals, Kubernetes, and container runtimes.
Why can't I use Git for my data pipelines?
So, if Pachyderm is similar to Git, why can't I store everything in Git rather than have multiple tools that I have to learn and support?, you might ask.
While Git is a great open source technology for software engineers, it is not tailored to solve the reproducibility problem in data science. This is mainly because Git was designed to track versions of files rather than establishing connections between data, code, and parameters – the essential parts of each data model.
While you can keep your code files in Git, doing so with the training data is not possible for the following reasons:
- File format support: Git is great for source code, such as text files, scripts, and so on, but it does not version non-binary files, such as image or video files. Git will detect that a binary file has changed, but it does not provide detailed information about the changes like it does in the case of text files. In addition, Git replaces the whole binary file, which makes for a dramatic increase in your repository when you update it.
- File size support: Often, image and video files are very large, and Git has a file size limitation of 100 MB per file. For best performance, Git recommends keeping your repository size below 1 GB, which might not be enough for some machine learning and deep learning projects.
- Cloning a large Git repository and all the associated history of each file can be problematic and takes a long time.
Consider these limitations when you plan the tooling for your machine learning project.
In addition to standard Git repositories, Git offers storage for large files, called Git Large File Storage (LFS). However, this option likely won't provide the needed functionality either. Git LFS has the following disadvantages for machine learning projects:
- Git LFS requires a server. You either need to set up and maintain your own Git LFS server or store it on a third-party online platform. Hosting your own server is problematic and requires additional support, and your preferred cloud provider might not be supported.
- Online Git LFS servers have a limited free tier. For projects that use high-resolution images or videos, this likely means that the limit will be reached fairly soon, and an upgrade to a paid version will be required.
- Designed for marketing department purposes, Git LFS seems like a good solution for marketing departments to store assets. Everyone else seems to prefer standard Git for their non-machine-learning projects.
Now that you know why source code version control systems are not the best solutions to work with data, let's review Pachyderm architecture.
Pachyderm architecture diagram
The following diagram shows the Pachyderm architecture:
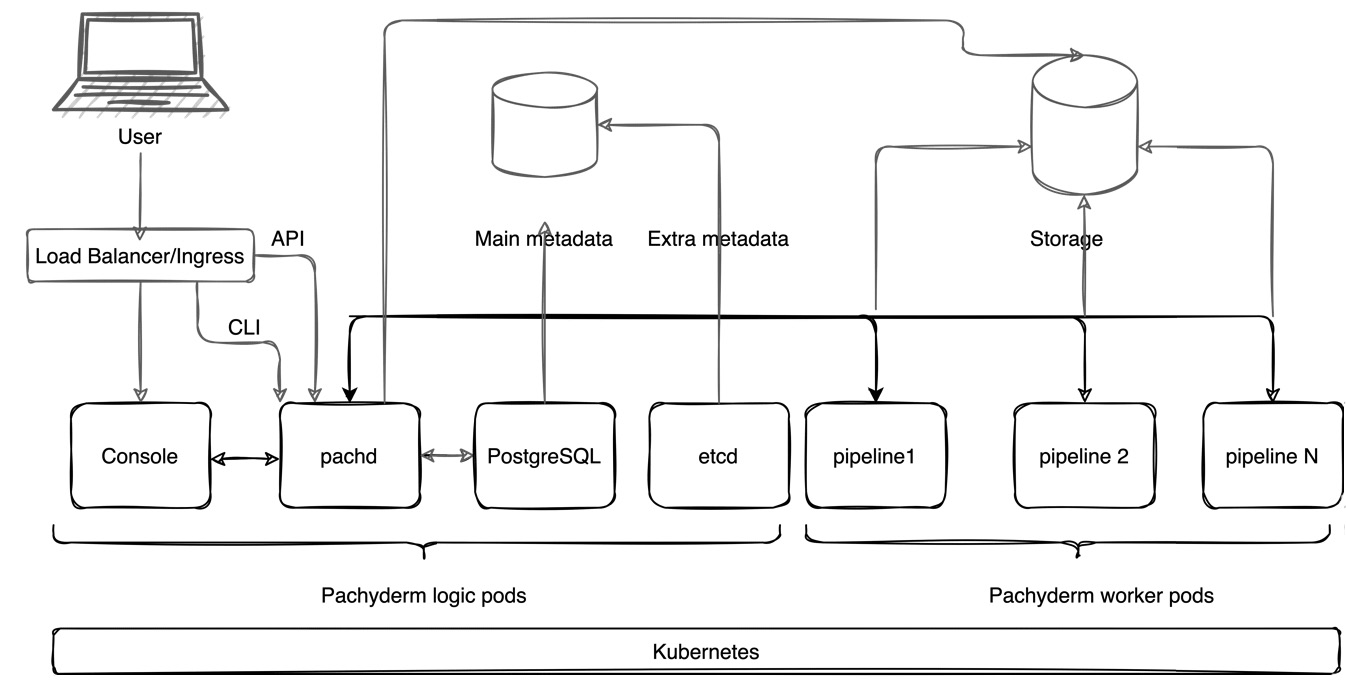
Figure 2.1 – Pachyderm architecture
Let's review all the components of this diagram one by one.
Kubernetes
Kubernetes, an open source distributed workflow scheduling system, is at Pachyderm's core. If you are not familiar with Kubernetes, here is a quick overview.
Kubernetes is the latest stage of infrastructure evolution, which started with traditional hardware deployments where data center operators deployed all applications on one physical server. The applications would fight over resources, which resulted in poor performance in some applications. This didn't scale well and was difficult to manage. Later on, we saw the rise of virtual machine technology that made the management of physical servers much easier and allowed us to run multiple applications on one physical server with better scalability. Many data centers still run a lot of virtual machines. Containers, the technology on which Kubernetes is based, made the infrastructure even leaner by allowing containers to share common operating system components and removing the redundant copies of the same software parts. Containers can be easily ported from one cloud platform to another, provide better resource allocation, and overall are easier to manage and support.
The function of Kubernetes is to manage containers and collections of containers called Pods. Today, Kubernetes is the dominant container technology in the open source world. Big tech companies might have their own types of schedulers – for example, Google has a project called Borg, but the majority of others are using one or another version of Kubernetes.
Companies such as Amazon, Microsoft, and Google offer hosted versions of Kubernetes called, respectively, AWS Elastic Kubernetes Service (EKS), Microsoft Azure Kubernetes Service (AKS), and Google Kubernetes Engine (GKE). Kubernetes can also be installed on-premises by using an automated software provisioning tool, such as Ansible, or using one of the specifically designed Kubernetes installation tools, such as Kubespray.
Some companies, such as DigitalOcean, provide a Kubernetes installation and management system that can deploy Kubernetes to a cloud platform of your choice.
Now that we know what Kubernetes is, let's review how Pachyderm leverages Kubernetes to provide a scalable, reproducible data science solution.
Helm
The tool that deploys Pachyderm on top of Kubernetes is called Helm. Helm is an open source package manager that enables you to deploy any Kubernetes-compatible packages on a Kubernetes cluster. The main advantage of Helm is that it makes the deployment of packages easy and allows a lot of flexibility through Helm Charts. A Helm Chart is a YAML file where you specify parameters for your Pachyderm cluster. Pachyderm provides sample Helm Charts for all types of installations, including local and cloud deployments. You can find an example of the values.yaml that you can use with the Pachyderm Helm Chart for local Pachyderm installation at https://github.com/pachyderm/pachyderm/blob/master/doc/docs/master/reference/helm_values.md.
In the values.yaml file, you can define everything you want to deploy with your Pachyderm cluster. For example, you can deploy the Pachyderm UI called Console, or not; you can configure storage buckets in your cloud provider; you can specify which version of Pachyderm to install, and so on.
Helm Charts are stored in Artifact Hub, located at https://artifacthub.io/. You can find Pachyderm at https://artifacthub.io/packages/helm/pachyderm/pachyderm. We will discuss the Pachyderm Helm Chart in more detail in Chapter 5, Installing Pachyderm on a Cloud Platform.
Pachyderm internals
When you deploy Pachyderm locally or on a cloud platform of your choice, the underlying Kubernetes orchestrator deploys the following components:
- Pachyderm Console (UI): The Pachyderm in-browser User Interface (UI) that provides basic Pachyderm functionality.
- Pachd: The Pachyderm daemon container that is responsible for managing all the main Pachyderm logical operations.
- postgress: Pachyderm requires a PostgreSQL instance to store metadata. PostgreSQL is a relational database popular in many open source and commercial solutions. While for testing purposes, an instance of PostgreSQL is shipped with the Pachyderm build, for production deployments and workloads, a separate cloud-based instance is strongly recommended.
- etcd: In older versions of Pachyderm, etcd was the key-value store that stored information about nodes and administrative metadata. In versions of Pachyderm 2.0.0 and later, etcd only stores a small portion of metadata, while the majority is stored in PostgreSQL.
Each of these components is deployed as a Kubernetes Pod—the smallest deployment unit of Kubernetes, which can have one or a collection of containers. Unlike in other applications, in Pachyderm, each Pod has only one container.
In addition to these containers that are deployed during the installation, every Pachyderm pipeline, a computational component that trains your model, is deployed as a separate Kubernetes Pod. These Pods are called workers. You can deploy an unlimited number of pipelines in your Pachyderm cluster and assign a custom amount of resources to each Pod. Pods are completely isolated from each other.
Other components
Figure 2.1 has the following components that we have not yet listed:
- Load Balancer or Ingress Controller: A networking component of Kubernetes that exposes HTTP/HTTPS routes from the cluster to the outside world. By default, Kubernetes provides a ClusterIP service that enables the services inside the cluster to talk to each other. There is no way to access cluster services from the outside world. In a production deployment, such as on a cloud platform, you need to deploy either an ingress controller or a load balancer to be able to access your Pachyderm cluster from the internet. In a local, testing deployment, Pachyderm uses port-forwarding. NodePort is another way to configure external access. However, it is not recommended for production deployments and, therefore, has been omitted from the current description. Pachyderm provides a Traefik ingress option to install through its Helm Chart.
- Pachyderm CLI or pachctl: pachctl is Pachyderm's Command-Line Interface (CLI), which enables users to perform advanced operations with Pachyderm pipelines and configure infrastructure as needed.
- API: Pachyderm supports programmatical access through a variety of language clients, including Python, Go, and others. Many users will find it useful to use one of the supported clients or even build their own.
- Metadata storage: etcd collects administrative metadata, which has to be stored either on a local disk or a Kubernetes Persistent Volume (PV), which can be on a cloud platform or any other platform of your choice.
- Object Storage: A location to store your Pachyderm File System (PFS) and the pipeline-related files. Pachyderm supports S3 object storage, such as MinIO, Google S3, Azure Blob storage, and Amazon S3.
In Figure 2.1, the user accesses the Pachyderm cluster through a load balancer or ingress controller by using either the Pachyderm dashboard, pachctl, or through the API by using a language client. The user's call is sent to pachd. pachd processes the request and updates the state of the pipeline and etcd accordingly. After the pipeline runs, it outputs the result to the configured storage location, from where it can be accessed by other third-party applications.
Container runtimes
A container runtime is software that executes the code in a container. There are many open source container runtimes, such as Docker, container, CRI-O, and others. Pachyderm supports the most popular container runtime—Docker. The fact that Pachyderm supports only one type of container runtime does not mean limited functionality. Most users will find Docker containers sufficient for all their needs.
To run a container, you need to have a container image and a place to store it. A container image is a file that contains the immutable code of a program that can be executed by the container runtime. Container images are stored in container registries, storage or repositories that host container images. Some container registries are public and others are private. All Pachyderm components are packaged as container images and stored in Docker Hub, a public container registry, meaning that anyone can download those images free of charge.
All your Pachyderm pipelines will have to be packaged as Docker containers and stored in a container registry. Your organization might require you to store your packages in a private container registry to protect the intellectual property of your pipeline. But if you are working on an open source or student project, you can store your container images in Docker Hub for free.
Now that we have reviewed the Pachyderm architecture, let's look at Pachyderm version control primitives that are foundational in understanding how Pachyderm works.
Learning about version control primitives
As we saw in a previous section, Pachyderm bears similarities to the code version control software called Git. If you have participated in developing an open source project before, you are likely familiar with Git through the use of a hosted Git version, such as GitHub, GitLab, or Gerrit.
Like with Git, you store your data in repositories, upload your data with a commit, and can have multiple branches in your repositories. Pachyderm stores the history of your commits and allows you to track changes or the history of your data back to its origins.
Pachyderm version control primitives enable you to go back in time and run your pipeline against previous versions of your changes. This can be very powerful in tracking bias and mistakes that crawl into your pipeline changes.
Let's look at these concepts in more detail.
Repository
A Pachyderm repository is a filesystem in which you store your data and where you store versions of your data. Pachyderm distinguishes between the input repository and output repository.
An input repository is a filesystem that you create and where you upload your data for further processing either by using a CLI, the UI, or automatically by using the API.
An output repository is a filesystem that Pachyderm creates automatically, which has the same name as the pipeline. This is the location where Pachyderm outputs the results of the calculation and from where results can be exported for serving as a model.
An important distinction from the Git repositories is that Pachyderm repository history is stored in a centralized location, which eliminates the risk of merge conflicts. Therefore, there is no equivalent of the .git history file in Pachyderm.
The following diagram demonstrates how internal and external repositories work within a pipeline:
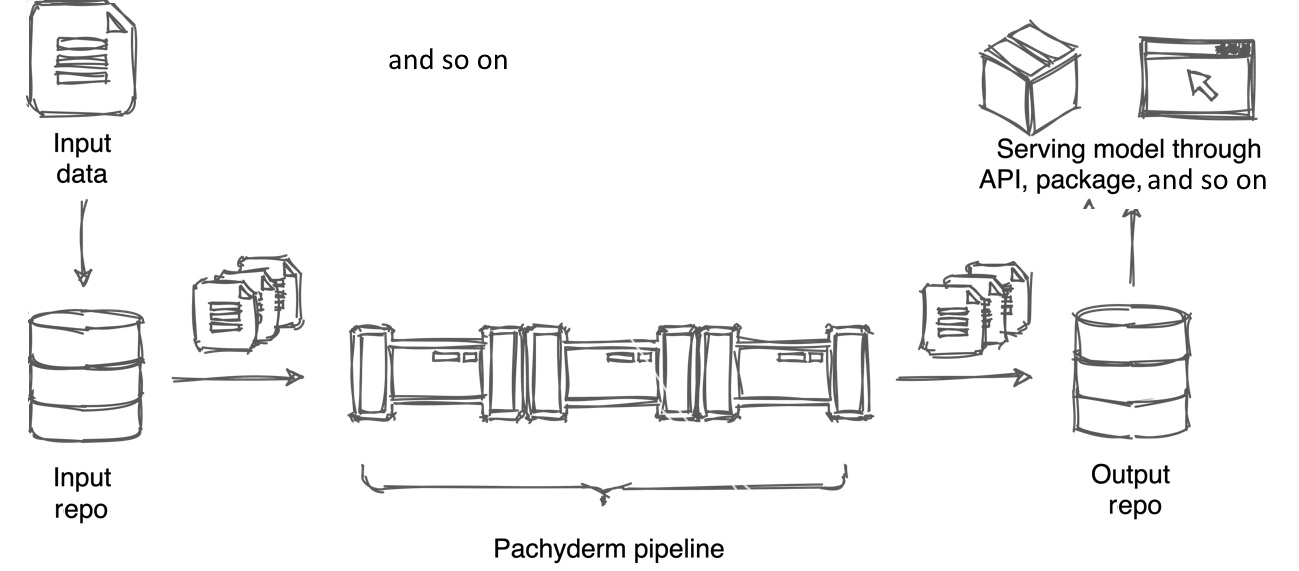
Figure 2.2 – Pachyderm input and output repositories
The preceding diagram shows a simple use case with just one pipeline. However, your workflow could be much more complex with multiple pipelines being intertwined and working together to achieve the needed result.
Branch
A branch is a Pachyderm line of development that tracks a set of changes within a repository. By default, a repository has no branches. The master branch that we will use in this book is just an example of how you can name your primary branch in Pachyderm, and it's not enforced. Typically, you create a branch on a repository when you upload initial data in the repository. You can create multiple branches to organize your work, but you likely won't use them as extensively as in Git. Often, just one branch is enough for all the work. You could create a separate branch for a different experiment. All branches are visible to all users, so you won't be able to have a local branch, experiment in it, and then merge it with the master branch as you do in Git.
To create a branch, run the following:
pachctl create branch <repo>@<branch>
The repository must exist before you can create a branch.
To confirm that a branch was created, run the following:
pachctl list branch <repo>
The final part of this section will deal with the commit concept.
Commit
A commit is a single immutable changeset of your data. For example, when new data is received from a streaming platform, such as Kafka, it is written to the Pachyderm repository as a new commit with an individual hash identifier.
The following diagram illustrates the concept of the commit on a branch in Pachyderm:
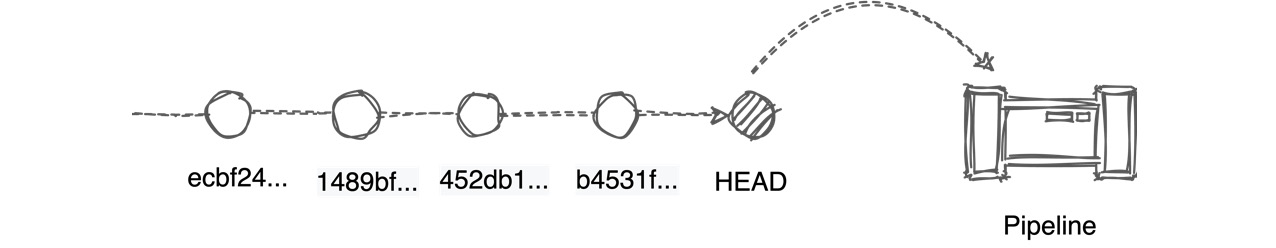
Figure 2.3 – A diagram showing Pachyderm commits
Data changes are stored in the repository history. The HEAD, or the HEAD commit of the branch, moves every time a new commit is submitted to the repository. As soon as the new data is submitted, the pipeline runs the code against these latest changes unless a different behavior is explicitly configured.
Now that we know the most important Pachyderm version control primitives, let's learn about the Pachyderm pipeline in more detail.
Discovering pipeline elements
This section walks you through the main Pachyderm pipeline concepts. The Pachyderm Pipeline System (PPS) is the centerpiece of Pachyderm functionality.
A Pachyderm pipeline is a sequence of computational tasks that data undergoes before it outputs the final result. For example, it could be a series of image processing tasks, such as labeling each image or applying a photo filter. Or it could be a comparison between two datasets or a finding similarities task.
A pipeline performs the following three steps:
- Downloads the data from a specified location.
- Applies the transformation steps specified by your cod.
- Outputs the result to a specified location.
The following diagram shows how a Pachyderm pipeline works:
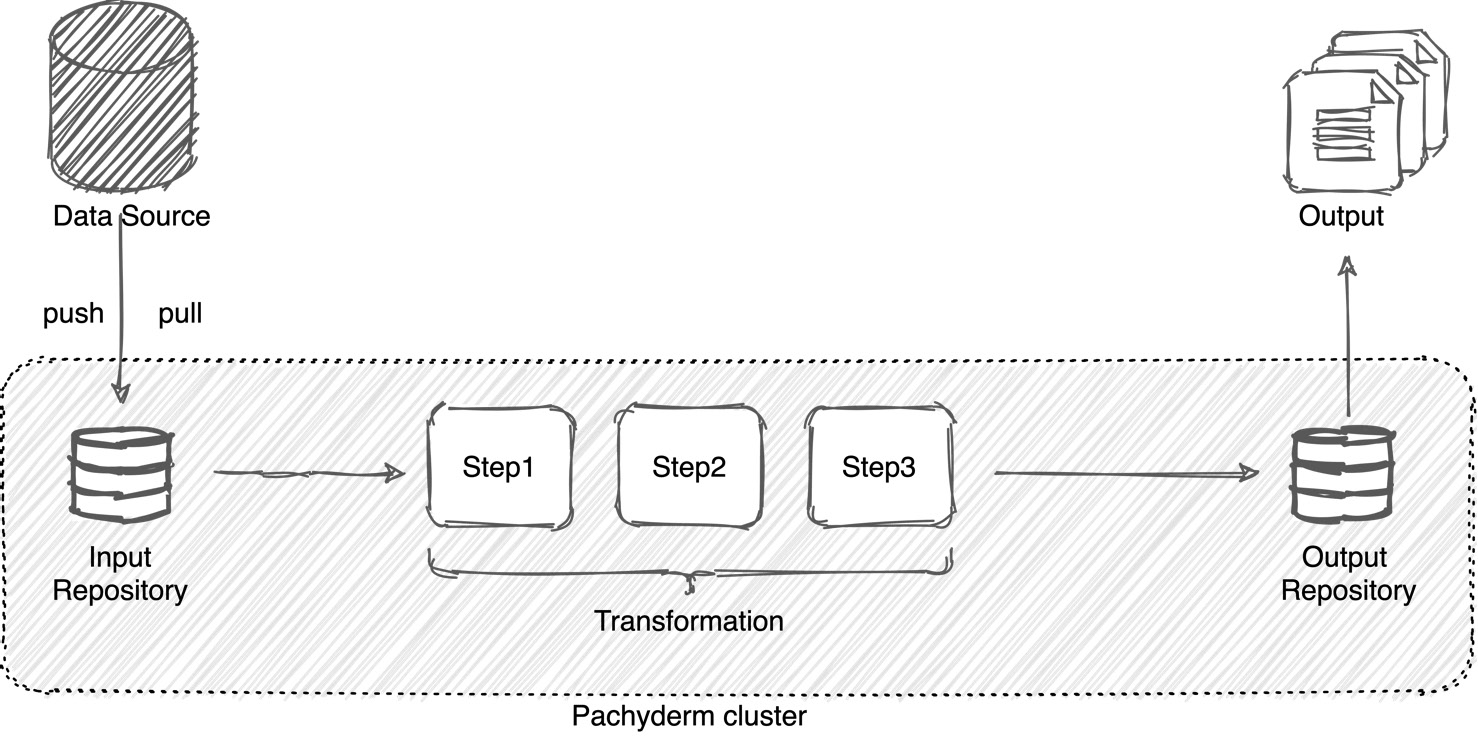
Figure 2.4 – Pachyderm pipeline
Each Pachyderm pipeline has an input and output repository. An input repository is a filesystem within Pachyderm where it is being placed from an outside source. This is the location inside of the pipeline Pod under the /pfs directory. The data can either be pulled by a pipeline or pushed to the input repository by the data source system. After the data goes through the transformation, it is placed into the output Pachyderm repository, which is located in the /pfs/out directory. From the output repository, the results can be further consumed by third-party applications or other pipelines.
Every time new data lands in the input repository, Pachyderm starts a pipeline job. The job processes the newly arrived data and places the results in the output repository.
Types of pipelines
A machine learning pipeline continuously performs the tasks that the data scientists coded on the new data. For example, you might want to compare or join two types of files together or apply certain parameters to them. To simplify these tasks, Pachyderm offers predefined pipelines that can do those things automatically.
Pachyderm offers the following types of pipelines:
- Standard pipeline
- Cron pipeline
- Spout pipeline
- Service pipeline
To create a pipeline, you need to write a pipeline specification. A pipeline specification is a file in the YAML Ain't Markup Language (YAML) or JavaScript Object Notation (JSON) format that describes what the pipeline needs to do. We will talk about pipeline specifications in more detail in the next chapter.
Standard pipelines
A standard pipeline, or simply a pipeline, is the most straightforward way to schedule work in a Pachyderm cluster. This type of pipeline is triggered when new data lands in the Pachyderm input repository. The pipeline spawns or resumes a Kubernetes Pod to run your code against the newly landed data and outputs the result to the output repository. The output repository is created automatically and has the same name as the pipeline.
The simplest standard pipeline must have the following components:
- name: A descriptive name for your pipeline. It is good practice to give your pipeline a name that reflects the task it accomplishes. For example, if your code analyzes the sentiment of users' feeds on social media, you might want to call it sentiment-analysis or something similar.
- transform: The transformation section of a pipeline contains the information about the Docker image that the pipeline needs to pull and use and the code that it needs to run in the pipeline container. For example, if you are doing sentiment analysis, you probably will use Natural Language Processing (NLP) tools such as Natural Language Toolkit (NLTK) or Stanford CoreNLP. Therefore, you can use either an already available Docker image for those tools or build your own custom one.
- input: An input repository from which the pipeline grabs data for processing. You will need to upload the data to an input repository either through the CLI, UI, or API.
The following text is an example of a pipeline specification that performs sentiment analysis. The pipeline is called sentiment-analyzer, uses the Docker image called ntlk-image, downloads data from the input repository called feed, and then runs the code stored in the file called transform_code.py:
# sentiment-analyzer.yml
---
pipeline:
name: sentiment-analyzer
transform:
image: nltk-image
cmd:
- python3
- "/transform_code.py"
input:
pfs:
repo: feed
glob: "/"
The glob parameter defines how to break down data into processable chunks that can be spread across multiple Pachyderm workers for better performance. We will discuss this parameter in more detail in the next section.
Important note
You can either put your transformation code in a file like in the example above or specify it directly in the cmd field. See Chapter 3, Pachyderm Pipeline Specification, for more information.
Cron pipelines
A Cron pipeline, or cron, is a pipeline that runs a specific task periodically. If you are familiar with the UNIX software utility cron, the Cron pipeline uses a similar logic—it runs periodically according to the specified schedule and performs the same task every time. Unlike the standard pipeline that runs every time new data arrives in the input repository, the Cron pipeline runs according to a schedule. You might want to use this pipeline to periodically scrape a website or a table.
A Cron pipeline specification would look very similar to the standard pipeline apart from the input section. You need to specify the time interval in the input section for your pipeline.
The following text is an example of a Cron pipeline in YAML format that scrapes a website called my-website once a day at midnight:
# scrape-my-website.yml
---
pipeline:
name: scrape-my-website
transform:
image: web-scraper
cmd:
- python3
- "/scraper.py"
input:
cron:
repo: mywebsite
spec: "@daily"
All the information about the website URL will go into your scraper.py file.
Spout pipelines
A spout pipeline, or spout, as the name suggests, is designed to stream data from an outside source, such as a publish/subscribe messaging system. In your infrastructure, you might have multiple message queue systems. With a Pachyderm spout, you can consolidate inputs from all of these systems and inject them into a Pachyderm spout pipeline. The spout code is not triggered by any event; instead, it runs continuously, listening for new messages.
Unlike in a standard or a Cron pipeline, spouts do not have an input repository, but instead, they listen on a specified address. The port and host can be specified either as an env variable in the pipeline specification or inside of the container.
Often, a spout is used together with a Pachyderm Service pipeline to expose the results of the pipeline because the data in the spout's output repository cannot be accessed with standard Pachyderm commands.
The following text is an example of a spout pipeline in YAML format that connects to an Amazon Simple Queue Service (SQS) host and applies a Python script, myscript.py, to the messages:
---
pipeline:
name: spout-pipeline
spout: {}
transform:
cmd:
- python3
- myscript.py
image: 'myimage'
env:
HOST: sqs-host
TOPIC: mytopic
PORT: '1111'
The spout section is left empty, but this is the place where you can combine the spout with a service pipeline to expose the results of the pipeline to the outside world.
Service pipelines
A service pipeline, or service, is a pipeline that can expose your output repository to the outside world. Unlike other pipelines, it does not perform any modifications to your data. The only function of this pipeline is to serve the results for your pipeline as an API, for example, in the form of a dashboard. A service pipeline is often combined with a spout pipeline and is sometimes called spout-service. However, it can use the results from any other pipeline output repository as its own input repository to expose the results.
A pipeline specification for this pipeline misses the transformation section. The following text is an example of a service pipeline in YAML format:
---
pipeline:
name: expose-service
input:
pfs:
glob: "/"
repo: final-results
service:
external_port: 31111
internal_port: 8888
Next, we'll see how Pachyderm works with datum.
Datum
A datum is the smallest piece of data that a Pachyderm worker can process. Folders and files in your input repository can be processed all as one datum or as multiple datums. This behavior is controlled by the glob parameter in the pipeline specification. Pachyderm processes each datum independently. If you have more than one worker running your pipeline, Pachyderm can schedule datums to run on separate workers for faster processing and, in the end, all datums are merged back together in the output repository.
Datums
This is the term used by the Pachyderm folks. Yes, usually, the plural is data, but datums is how it appears throughout Pachyderm's documentation!
Breaking down your data into multiple datums provides the following advantages:
- Improves performance by scaling your pipeline to multiple Pachyderm workers
- Enables you to process only specific files and folders
For example, say you have a repository with the following folder structure:
data
├── folder1
│ ├── file1
│ ├── file2
│ └── file3
├── folder2
│ ├── file1
│ └── subfolder1
│ └── file1
└── folder3
├── subfolder1
│ ├── file1
│ └── file2
└── subfolder2
├── file1
└── file2
For this folder structure, you can set the following types of glob patterns:
- /: Process all files and folders as a single datum. This single datum includes all files and folders in the / directory.
- /*: Process each folder as a separate datum. The resulting number of datums is three, including the following:
/folder1
/folder2
/folder3
- /*/*: Process each filesystem object on the /*/* level as a separate datum. The resulting number of datums is seven, including the following:
/folder1/file1
/folder1/file2
/folder1/file3
/folder2/file1
/folder2/subfolder1
/folder3/subfolder1
/folder3/subfolder2
- /*/*/*: Process each filesystem on the third level. The resulting number of datums is five, including the following:
/folder2/subfolder1/file1
/folder3/subfolder1/file1
/folder3/subfolder1/file2
/folder3/subfolder2/file1
/folder3/subfolder2/file2
- /**: Process each file, folder, and subfolder as a separate datum. The resulting number is 15, which includes all files and folders listed in the preceding code extract:
/folder1/
/folder1/file1
/folder1/file2
/folder1/file3
/folder2/
/folder2/file1/
/folder2/subfolder1/
...
- /<filesystem>/*: Process only the files and folders that match the naming pattern. For example, if you decide to process only the data in folder1, you will set your glob pattern to /folder1/*. Similarly, you can set just the first few letters of a directory name as a glob pattern.
These are the most commonly used glob patterns. Extended glob patterns are available with the Pachyderm's ohmyglob library. For more information, see https://github.com/pachyderm/ohmyglob.
Scaling your pipeline with datums
One of the biggest advantages of breaking your data into multiple datums is the ability to scale your work across multiple pipeline workers to significantly improve the performance and the processing time of your pipeline. By default, Pachyderm deploys one worker node for each pipeline, but you can increase this number as needed by specifying the parallelism_spec parameter in the pipeline specification.
The following diagram demonstrates how datums are scaled across multiple workers:
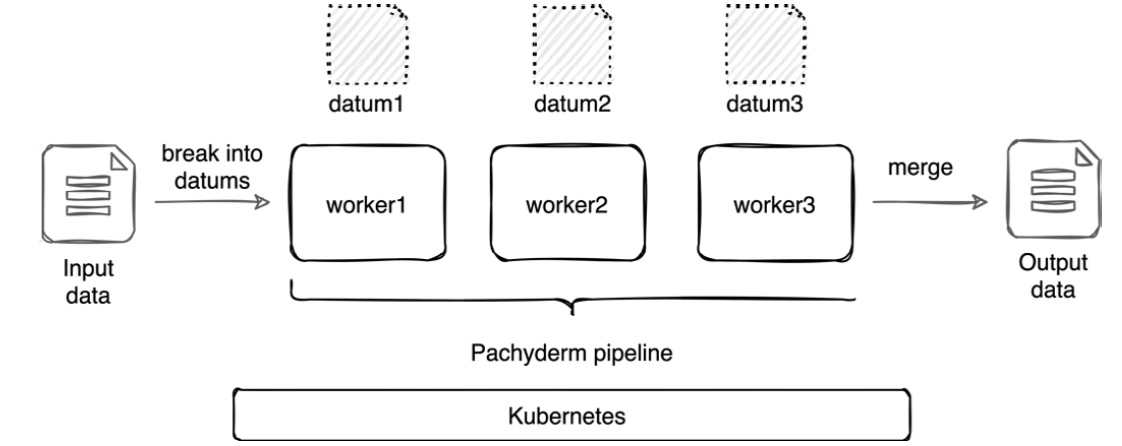
Figure 2.5 – Scaling a pipeline
In the preceding diagram, the input data is broken down into three workers, which simultaneously start the processing. After all of the datums are processed, they are merged into the final output result. It is important to emphasize that datums only exist inside of a Pachyderm pipeline. They cannot be accessed, mounted, or modified in any way.
Pachyderm inputs
In Pachyderm, you likely will create multiple repositories to store training data, testing data, and parameters. Pachyderm provides an automated way to combine files in different repositories in the pipeline to be processed together through the use of inputs. If you have used SQL before, you might find that these types of inputs remind you of SQL operators. However, there is one major difference that distinguishes SQL operators from Pachyderm inputs. While in SQL you can create matching pairs of rows in each table, Pachyderm inputs work on the file level only. This means that you can only combine files or directories inside Pachyderm repositories based on their names rather than the contents of the files.
Important note
This section provides examples of how you can create various Pachyderm inputs. These examples are for your reference only and you might want to use some of them in your future Pachyderm work after you install and configure Pachyderm. We will cover the installation and setup in the Installing Pachyderm locally and Deploying Pachyderm on a cloud platform sections of Chapter 3, Pachyderm Pipeline Specification.
Cross inputs
A cross input, or cross, is an input in which each file from one repository is combined with a file in the other repository. The set of matched files is determined by the glob pattern, and all that data is visible to the pipeline code at the time of the pipeline run.
The input part of your pipeline file in YAML format might look like this:
…
input:
cross:
- pfs:
glob: "/*"
repo: data
- pfs:
glob: "/"
repo: parameters
…
For example, you have two Pachyderm repositories, data and parameters, with the following structure and files:
data
├── image1.png
├── image2.png
├── image3.png
└── image4.png
parameters
├── param1.csv
└── param2.csv
If you create a pipeline that creates a cross-product of these repositories with a glob pattern set to /*, you will get eight datums in total—each file from the data repository will be combined with each file in the parameters repository. This is how the files will be processed:
data@4e5897...:/image1.png, parameters@d1a0da...:/param1.csv
data@4e5897...:/image2.png, parameters@d1a0da...:/param1.csv
data@4e5897...:/image3.png, parameters@d1a0da...:/param1.csv
data@4e5897...:/image4.png, parameters@d1a0da...:/param1.csv
data@4e5897...:/image1.png, parameters@d1a0da...:/param2.csv
data@4e5897...:/image2.png, parameters@d1a0da...:/param2.csv
data@4e5897...:/image3.png, parameters@d1a0da...:/param2.csv
data@4e5897...:/image4.png, parameters@d1a0da...:/param2.csv
In the preceding output, each pair of files and each line is one datum.
Union inputs
A union input, or union, is an input that enables you to combine datums from one repository with datums in another repository. The total number of datums is the sum of datums in all repositories. If we take the same repository structure as described in the Cross inputs section and, instead of creating a cross-product, just add them to each other by using the union input, we will have a total of five datums.
The input part of your pipeline file in YAML format might look like this:
…
input:
union:
- pfs:
glob: "/*"
repo: data
- pfs:
glob: "/"
repo: parameters
…
The following output demonstrates the list of datums for a pipeline with the /* glob pattern set for both repositories:
data@4e58978ca1304f16a0a5dfe3715363b4:/image1.png
data@4e58978ca1304f16a0a5dfe3715363b4:/image2.png
data@4e58978ca1304f16a0a5dfe3715363b4:/image3.png
data@4e58978ca1304f16a0a5dfe3715363b4:/image4.png
model@1de402b7be004a289f6d7185b2329b21:/
Your code processes each file individually and recognizes only one file with every run, ignoring all other files.
Join inputs
A join input, or join, is a Pachyderm input that enables you to match pairs of files based on a specific naming pattern defined by a capturing group. A capturing group is a regular expression that is defined within parentheses. The capturing group must be specified in the join_on parameter in your Pachyderm pipeline.
Pachyderm separates inner joins and outer joins. The difference between the two is that an inner join matches pairs of files, skipping files that do not match the specified pattern. An outer join acts the same way as an inner join, but also includes files that do not match the pattern in the pipeline run. If this sounds a bit confusing, the following example should clarify how the pipeline works.
Let's say you have two repositories, data and parameters, and the parameters repo has the following structure:
NAME TYPE SIZE
/param-0101-2021.txt file 4B
/param-0102-2021.txt file 4B
/param-0103-2021.txt file 4B
/param-0104-2021.txt file 4B
/param-0105-2021.txt file 4B
The data repository has the following structure:
NAME TYPE SIZE
/data-0101-2021.txt file 2B
/data-0102-2021.txt file 2B
/data-0103-2021.txt file 2B
/data-0104-2021.txt file 2B
/data-0105-2021.txt file 2B
/data-0106-2021.txt file 2B
/data-0107-2021.txt file 2B
In your pipeline, you might want to match the files by their date. To do so, you would have to specify a capturing group of $1 and a glob pattern, /. Here is an example of a pipeline specification in YAML format that matches these file paths:
---
pipeline:
name: describe
input:
join:
- pfs:
glob: "/data-(*).txt"
repo: data
join_on: "$1"
- pfs:
glob: "/param-(*).txt"
repo: parameters
join_on: "$1"
transform:
image: mydockerhub/describe
cmd:
- python3
- "/describe.py"
This capturing group in combination with the / glob pattern matches five pairs of files:
data@2c95b1...:/data-0101-2021.txt, parameters@b7acec...:/param-0101-2021.txt
data@2c95b1...:/data-0102-2021.txt, parameters@b7acec...:/param-0102-2021.txt
data@2c95b1...:/data-0103-2021.txt, parameters@b7acec...:/param-0103-2021.txt
data@2c95b1...:/data-0104-2021.txt, parameters@b7acec...:/param-0104-2021.txt
data@2c95b1...:/data-0105-2021.txt, parameters@b7acec...:/param-0105-2021.txt
/data-0106-2021.txt and /data-0107-2021.txt do not have matching pairs, and therefore, Pachyderm would skip them in this run. However, if you want to include these in the files in the pipeline run, you can specify outer_join: true in the data repository input to include these files in the pipeline run without a pair. This abstract shows how you can add this parameter:
…
input:
join:
- pfs:
glob: "/data-(*).txt"
repo: data
join_on: "$1"
outer_join: true
…
Then the list of datums in your pipeline will look like this:
data@2c95b1...:/data-0101-2021.txt, parameters@b7acec...:/param-0101-2021.txt
data@2c95b1...:/data-0102-2021.txt, parameters@b7acec...:/param-0102-2021.txt
data@2c95b1...:/data-0103-2021.txt, parameters@b7acec...:/param-0103-2021.txt
data@2c95b1...:/data-0104-2021.txt, parameters@b7acec...:/param-0104-2021.txt
data@2c95b1...:/data-0105-2021.txt, parameters@b7acec...:/param-0105-2021.txt
data@2c95b1...:/data-0106-2021.txt
data@2c95b1...:/data-0107-2021.txt
data-0106-2021.txt and data-0107-2021 are included in the pipeline run without a pair.
Group inputs
A group input, or group, is a Pachyderm input that enables you to group matching files within one or more repositories based on a specific naming pattern. This type of input is similar to joins but it can address a broader use case. In a group input, you must specify a replacement group under the group_by pipeline parameter. If we took the same pipeline that we used in the inner join input example and replaced join with group and join_on with group_by, we would get the exact same result as we got with the inner join input.
The main differences from the join input include the following:
- A group input creates one large datum of the files that match a naming pattern, while a join crosses two files that match a pattern and typically creates more datums.
- A group input enables you to match pairs in a single repository, while a join requires at least two repositories.
- A group input is primarily useful in time series scenarios when you need to group files based on their timestamps.
Therefore, you could create a cron pipeline with the replacement group that matches a specific timestamp.
For example, say you have repository data with the following structure:
/data-0101-2020.txt file 2B
/data-0101-2021.txt file 2B
/data-0102-2020.txt file 2B
/data-0102-2021.txt file 2B
/data-0103-2020.txt file 2B
/data-0103-2021.txt file 2B
/data-0104-2021.txt file 2B
/data-0105-2021.txt file 2B
/data-0106-2021.txt file 2B
/data-0107-2021.txt file 2B
Your pipeline in YAML format might look like this:
---
pipeline:
name: test-group
transform:
cmd:
- python3
- "/test.py"
input:
group:
- pfs:
repo: data
glob: "/data-(*)-(*).txt"
group_by: "$1"
You have two replacement groups listed in the pipeline. Replacement group one matches the month and day in the name of the text file. For example, in the file /data-0104-2021.txt, it is 0104. The second replacement group matches the year in the timestamp. In the same file, it is 2021.
If you specify the first matching group in your pipeline, the resulting list of datums will include three pairs and seven datums in total:
data@...:/data-0101-2020.txt, data@..:/data-0101-2021.txt
data@...:/data-0102-2020.txt, data@...:/data-0102-2021.txt
data@...:/data-0103-2020.txt, data@...:/data-0103-2021.txt
data@...:/data-0104-2021.txt
data@...:/data-0105-2021.txt
data@...:/data-0106-2021.txt
data@...:/data-0107-2021.txt
In the preceding output, the files with the matching day and month, such as 0101, are grouped in one datum. If you change the group_by parameter to use the second replacement group, meaning grouping by year, the list of datums will include two datums, grouping the files by year:
- data@ecbf241489bf452dbb4531f59d0948ea:/data-0101-2020.txt, data@ecbf241489bf452dbb4531f59d0948ea:/data-0102-2020.txt, data@ecbf241489bf452dbb4531f59d0948ea:/data-0103-2020.txt
- data@ecbf241489bf452dbb4531f59d0948ea:/data-0101-2021.txt, data@ecbf241489bf452dbb4531f59d0948ea:/data-0102-2021.txt, data@ecbf241489bf452dbb4531f59d0948ea:/data-0103-2021.txt, data@ecbf241489bf452dbb4531f59d0948ea:/data-0104-2021.txt, data@ecbf241489bf452dbb4531f59d0948ea:/data-0105-2021.txt, data@ecbf241489bf452dbb4531f59d0948ea:/data-0106-2021.txt, data@ecbf241489bf452dbb4531f59d0948ea:/data-0107-2021.txt
In this section, we have learned about the most important Pachyderm pipeline primitives and how they differ from each other.
Summary
In this chapter, we have learned about the most important Pachyderm version control primitives, including repositories, branches, and commits. We reviewed that although they are similar to those of the Git version control system, they have a few very important differences.
We've learned about the types of Pachyderm pipelines and inputs and how a pipeline can be scaled and optimized through the use of glob patterns and datums.
In the next chapter, we will review the Pachyderm pipeline specification in more detail and will learn how to use the various pipeline settings to run a pipeline in the most efficient way.
Further reading
- Git documentation: https://git-scm.com/
- Kubernetes documentation: https://kubernetes.io/docs/home
- Helm documentation: https://helm.sh/docs/
