9k-Nearest neighbor algorithm
This chapter covers the following items:
–Algorithm for computing the distance between the neighboring data
–Examples and applications
The methods used for classification (Bayesian classification, decision tree induction, rule-based classification, support vector machines, classification by back propagation and classification based on association rule mining) mentioned up until this point in our book have been the examples of eager learners. By eager learners, we mean that there will be a construction of a generalization based on a set of training datasets prior to obtaining new datasets to be classified. The new datasets are also in the category of test. It is called eager because the learned model is ready and willing to do the classification of the datasets that have not been seen previously [1–4]. As opposed to a lazy approach where the learner waits until the last minute before it does any model of construction for the classification of a test dataset, lazy learner or learning from your neighbors just carries out a simple storing or very minor processing, and waits until given to a test dataset. Upon seeing the test dataset, it carries out generalization for the classification of the dataset depending on its similarity to the training datasets stored. When we make a contrast with eager learning methods, lazy learners as its name suggests do less work when a training dataset is presented but it is supposed to do more work when it does a numeric estimation or a classification. Since lazy learners store the training datasets, or “instances”, we also call them. instance-based learners. Lazy learners prove to be expensive in terms of computational issues while they are engaged in numeric estimation of a classification since they need efficient techniques for storage. They also present little explanation for the structure of the data. Another aspect of lazy learners is that they support incremental learning, capable of modeling complex decision spaces that have hyperpolygonal shapes that are not likely to be described easily through other learning algorithms [5].
One of the examples of lazy learners is k-nearest neighbor classifiers [6]. The first description of k-nearest neighbor method was made in the early 1950s. The method requires intensive work with large training sets. It gained popularity after the 1960s (this was the period when increased power of computation became accessible). As in the 1960s, they have been used extensively in pattern recognition field. Based on learning by analogy, nearest neighbor classifiers include a comparison of a particular test dataset with training datasets that are similar to it. The training datasets are depicted by n attributes. Each dataset represents a point in an n-dimensional space. Thus, all the training datasets are stored in n-dimensional pattern space. If there is a case of an unknown dataset, then a k-nearest neighbor classifier will look for a pattern space for the K training datasets, which will be nearest/closest to the unknown dataset [7–8]. These K training datasets are therefore the k “nearest neighbors” of the unknown dataset. We define closeness based on a distance metric similar to the Euclidean distance, which is between two points or datasets: X1 = (x11, x12, ..., x1n) and X2 = (x21, x22, ..., x2n) that gives the following formula:
We can explain this procedure as follows: the difference between the corresponding values of the attribute in dataset X1 and in dataset X2 is taken for each numeric attribute. The square of this difference is taken and accumulated. From the total accumulated distance count, the square root is taken. The values of each attribute is normalized before the use of eq. (9.1). Thus, this process allows the prevention of attributes that have initially large ranges (e.g., net domestic credit) from balancing attributes with initially smaller ranges (e.g., binary attributes). Min-max normalization can be utilized for transforming a value v of a numeric attribute A to v′ in the range [0, 1]:
minA and maxA are the minimum and maximum values of attribute A. In k-nearest neighbor classification, the most common class is allotted to the unknown dataset among its k-nearest neighbors. Nearest neighbor classifiers are also used for numerical prediction to return a real-valued prediction for a particular dataset that is unknown [9–10]. What about the distance to be calculated for attributes that are not numeric but categorical or nominal in nature? For such nominal attributes, comparing the corresponding value of the attribute in dataset with the one in dataset X2 is used as a simple method. If two of them are identical (e.g., datasets X1 and X2 have the EDSS score of 4), the difference between the two is considered and taken as 0. If the two are different (e.g., dataset X1 is 4. Yet, dataset X2 is 2), the difference is regarded and taken as 1. As for the missing values, generally the following is to be applied: the maximum possible difference is assumed if the value of a given attribute A is missing in dataset X1 and/ or dataset X2. The difference value is taken as 1 for nominal attributes if the corresponding values of A is missing or both of them are missing. In an alternative scenario, let us assume that A is numeric and missing from both X1 and X2 datasets, then the difference is also taken to be 1. If only one value is missing and the other one that is called v′ is existent and normalized, it is possible to take the difference as |1 − v′| or |0 − v′|(|1 − v′|or v′), depending on which one is higher.
Another concern at this point is about determining a good value for k (the number of neighbors). It is possible to do the determination experimentally. You start with k = 1, and use a test set in order to predict the classifier’s error rate. This is a process which you can repeat each time by incrementing k to permit one more neighbor. You may therefore select the k value that yields the minimum error rate. Generally, when the number of training datasets is large, then the value of k will also be large. Thereby, you base the prediction decisions pertaining to classification and numeric on a larger slice of the stored datasets. Distance-based comparisons are used by nearest neighbor classifiers. Distance-based comparisons essentially give equal weight to each of the attributes. For this reason, it is probable that they may have poor accuracy with irrelevant or noisy attributes. This method has been subjected to certain modification so that one can merge attribute weighting and the pruning of datasets with noisy data. Choosing a distance metric is a crucial decision to make. It is also possible that you use the Manhattan (city block) distance or other distance measurements. It is known that nearest neighbor classifiers are tremendously slow while they are carrying out the classification of test datasets. Let us say D is a training dataset of |D|datasets and k = 1, and O(|D|) comparisons are needed for the classification of a given test dataset. You can reduce the number of comparisons to O(log|D| by arranging the stored datasets into search trees. Doing the application in parallel fashion may allow the reduction of the running time to a constant: O(1) independent of |D|. There are some other techniques that can be employed for speeding the classification time. For example, you can make use of partial distance calculations and editing the stored datasets. In the former one, namely in partial distance method, you calculate the distance depending on a subset of the n attributes. In case this is beyond a threshold, then you halt the further computation for the prearranged stored dataset. This process goes on with the next stored dataset. As for the editing method, training datasets that end up being useless are removed. In a way, a clearing is made for the unworkable ones, which is also known as pruning or condensing because of the method reducing the total number of stored datasets [11–13].
The flowchart of k-nearest neighbor algorithm is provided in Figure 9.1.
Below you can find the steps of k-nearest neighbor algorithm (Figure 9.2).
According to this set of steps, each dataset signifies a point in an n-dimensional space. Thereby, all the training datasets are stored in an n-dimensional pattern space. For an unknown dataset, a k-nearest neighbor classifier explores the pattern space for the K training datasets that are closest to the unknown dataset with distance as shown in the second step of the flow. The K training datasets give us the k “nearest neighbors” of the unknown dataset.
In algorithm Figure 9.2, the step by step progress stages have been provided for the k-nearest neighbor algorithm. This concise summary will show the readers that it is simple to follow this algorithm as the steps will show.

Identifying the training and dataset to be classified, D training dataset that contains n number of samples is applied to k-nearest neighbor algorithm as the input. The attributes of each sample in D training set are represented as x1, x2, . . . , xn.
Compute distance dist(xi, xj): Each sample of the test dataset to be classified, the distance to all the samples in the training dataset is calculated according to eq. (9.1). Which training samples the smallest K unit belongs to is calculated among the distance vector obtained (argsort function denotes the ranking of the test sample according to the distance value regarding the samples) [3]. Take a look at the class label data of the k number of neighbors that are closest. Among the label data of the neighbors, the most recurring value is identified as the class of the x test data.

9.1k-Nearest algorithm for the analysis of various data
k nearest algorithm has been applied on economy (U.N.I.S.) dataset, multiple sclerosis (MS) dataset and WAIS-R dataset in Sections 9.1.1, 9.1.2 and 9.1.3, respectively.
9.1.1k-Nearest algorithm for the analysis of Economy (U.N.I.S.)
As the second set of data, we will use in the following sections, some data related to U.N.I.S. countries’ economies. The attributes of these countries’ economies are data regarding years, unemployed youth male(% of male labor force ages 15–24) (national estimate), gross value added at factorcost ($), tax revenue, gross domestic product (GDP) growth (annual %). Economy (U.N.I.S.) Dataset is made up of a total of 18 attributes. Data belonging to economies of countries such as USA, New Zealand, Italy and Sweden from 1960 to 2015 are defined on the basis of attributes given in Table 2.8 Economy (U.N.I.S.) Dataset, to be used in the following sections. For the classification of D matrix through k-nearest neighbor algorithm, the first-step training procedure is to be employed. For the training procedure, 66.66% of the D matrix can be split for the training dataset (151 × 18) and 33.33% as the test dataset (77 × 18).
How can we find the class of a test data belonging to Economy (U.N.I.S.) dataset provided in Table 2.8 according to k-nearest neighbor algorithm? We can a find the class label of a sample data belonging to Economy (U.N.I.S.) dataset through the following steps (Figure 9.3) specified below:

Step (1) Let us split the bank dataset as follows: 66.66% as the training data (D = 151 × 18) and 33.33% as the test data (T = 77 × 18).
Step (2–3) Let us calculate the distance of the data to be classified to all the samples in accordance with the Euclid distance (eq. 9.1)
D = (12000, 45000, ..., 54510) and T = (2000, 21500, ..., 49857), and the distance between the data is computed according to eq. (9.1).
Please bear in mind that our Economy dataset comprised 18 individual attributes and we see that it is (1 ≤ i ≤ 18)
Step (5–9) Let us find which class the smallest k number of neighbors belong to with regard to training samples among the distance vector obtained dist(D, T) = dist(D, T) = [dist(D, TÞ1, dist(D, TÞ2, ..., dist(D, T)18]
How can we find the class of a test data belonging to Economy data according to k-nearest neighbor algorithm?
We can find the class label of a sample data belonging to Economy dataset through the following steps specified below:
Step (1) The Economy dataset samples are positioned to a 2D coordinate system as provided below: the blue dots represent USA economy, the red ones represent New Zealand economy, the orange dots represent Italy economy, and finally the purple dots show Sweden economy classes.

Step (2) Assume that the test data that we have classified on the coordinate plane where Economy dataset exists corresponds to the green dot. The green dot selected from the dataset of Economy of USA, NewZealand, Italy, Sweden and data (whose class is not identified) has been obtained randomly from the dataset, which is the green dot.

Step (3) Let us calculate the distances of the new test data to the training data according to Euclid distance formula (eq. 9.1). By ranking the distances calculated, find the three nearest neighbors to the test data.
Step (4) Two of the three nearest members belong to purple dot class. Therefore, we can classify the class of our test data as purple (Sweden economy).

As a result, the data in the Economy dataset with 33.33% portion allocated for the test procedure and classified as USA (U.), New Zealand(N.), Italy(I.) and Sweden(S.) have been classified with an accuracy rate of 84.6% based on three nearest neighbor algorithm.
9.1.2k-Nearest algorithm for the analysis of multiple sclerosis
As presented in Table 2.12, multiple sclerosis dataset has data from the following groups: 76 samples belonging to RRMS, 76 samples to SPMS, 76 samples to PPMS and 76 samples belonging to healthy subjects of control group. The attributes of the control group are data regarding brain stem (MRI 1), corpus callosum periventricular (MRI 2), upper cervical (MRI 3) lesion diameter size (milimetric [mm]) in the MR image and EDSS score. Data are made up of a total of 112 attributes. It is known that using these attributes of 304 individuals the data if they belong to the MS subgroup or healthy group is known. How can we make the classification as to which MS patient belongs to which subgroup of MS (including healthy individuals and those diagnosed with MS (based on the lesion diameters (MRI 1,MRI 2,MRI 3), number of lesion size for MRI 1, MRI 2, MRI 3 as obtained from MRI images and EDSS scores?) D matrix has a dimension of (304 × 112). This means D matrix includes the MS dataset of 304 individuals along with their 112 attributes (see Table 2.12 for the MS dataset). For the classification of D matrix through Naive Bayesian algorithm the first-step training procedure is to be employed. For the training procedure, 66.66% of the D matrix can be split for the training dataset (203 × 112) and 33.33% as the test dataset (101 × 112).
Following the classification of the training dataset being trained with k-nearest algorithm, we can do the classification of the test dataset (Figure 9.4).

To be able to classify through k-nearest neighbor algorithm, the test data has been selected randomly from the MS dataset as 33.33%.
Let us now think for a while about how to classify the MS dataset provided in Table 2.12 in accordance with the steps of k-nearest neighbor algorithm (Figure 9.3).
Step (1) Let us identify 66.66% of the MS dataset as the training data (D = 203 × 112) and 33.33% as the test data (t = 101 × 112).
Step (2–3) Let us calculate the distance of the data to be classified to all the samples in accordance with the Euclid distance (eq. 9.1.). D = (1, 4, ..., 5.5) and T = (2, 0, ..., 7), and let us calculate the distance between the data according to eq. (9.1).
Please bear in mind that the MS dataset comprised 112 attributes (attribute) (1 ≤ i ≤ 120)
Step (5–9) Let us now find which class the smallest k number of neighbors belong to among the distance vector obtained dist(D, T) = [dist(D, T)1, dist(D, T)2, ..., dist(D, T)120].
How can we find the class of a test data belonging to the MS dataset according to k-nearest neighbor algorithm?
We can find the class label of a sample data belonging to MS dataset through the following steps specified below:
Step (1) The MS dataset samples are positioned to a 2D coordinate system as provided below: the blue dots represent PPMS, the red ones represent SPMS, the orange dots represent RRMS, and purple dots represent the healthy subjects class.

Step (2) Assume that the test data that we have classified on the coordinate plane where the MS dataset exists corresponds to the green dot. The green dot selected from the dataset of the MS data (PPMS, RRMS, SPMS) and data (whose class is not identified) has been gained randomly from the dataset.

Step (3) The distances of the subgroups in the other MS dataset whose selected class is not identified is calculated in accordance with Euclid distances. The Euclid distances of the test data to the training data are calculated. Let us find the three nearest neighbors by ranking the distances calculated from the smallest to the greatest (unclassified).

Step (4) The two of the three nearest neighbors belonging to the blue dot class have been identified according to Euclid distance formula. Based on this result, we identify the class of the test data as blue. The dot denoted in blue color can be classified as PPMS subgroup of MS.
As a result, the data in the MS dataset with 33.33% portion allocated for the test procedure and classified as RRMS, SPMS, PPMS and Healthy have been classified with an accuracy rate of 68.31% based on three nearest neighbor algorithm.
9.1.3k-Nearest algorithm for the analysis of mental functions
As presented in Table 2.19, the WAIS-R dataset has the following data: 200 belonging to patient and 200 samples belonging to healthy control group. The attributes of the control group are data regarding School Eeducation, Gender and D.M. Data are made up of a total of 21 attributes. It is known that using these attributes of 400 individuals, the data if they belong to patient or healthy group are known. How can we make the classification as to which individual belongs to which patient or healthy individuals and those diagnosed with WAIS-R test (based on the School Education, Gender and D.M)?D matrix has a dimension of 400 × 21. This means D matrix includes the WAIS-R dataset of 400 individuals along with their 21 attributes (see Table 2.19) for the WAIS-R dataset. For the classification of D matrix through k-nearest neighbor algorithm, the first-step training procedure is to be employed. For the training procedure, 66.66% of the D matrix can be split for the training dataset (267 × 21) and 33.33% as the test dataset (133 × 21). Following the classification of the training dataset being trained with k-nearest algorithm, we can classify the test dataset. Following the classification of the training dataset being trained with k-nearest algorithm, we can classify the test dataset.
How can we classify the WAIS-R dataset in Table 2.19 in accordance with the steps of k-nearest neighbor algorithm (Figure 9.5)?
Step (1) Let us identify the 66.66% of WAIS-R dataset as the training data (D = 267 × 21) and 33.33% as the test data (k = 133 × 21).
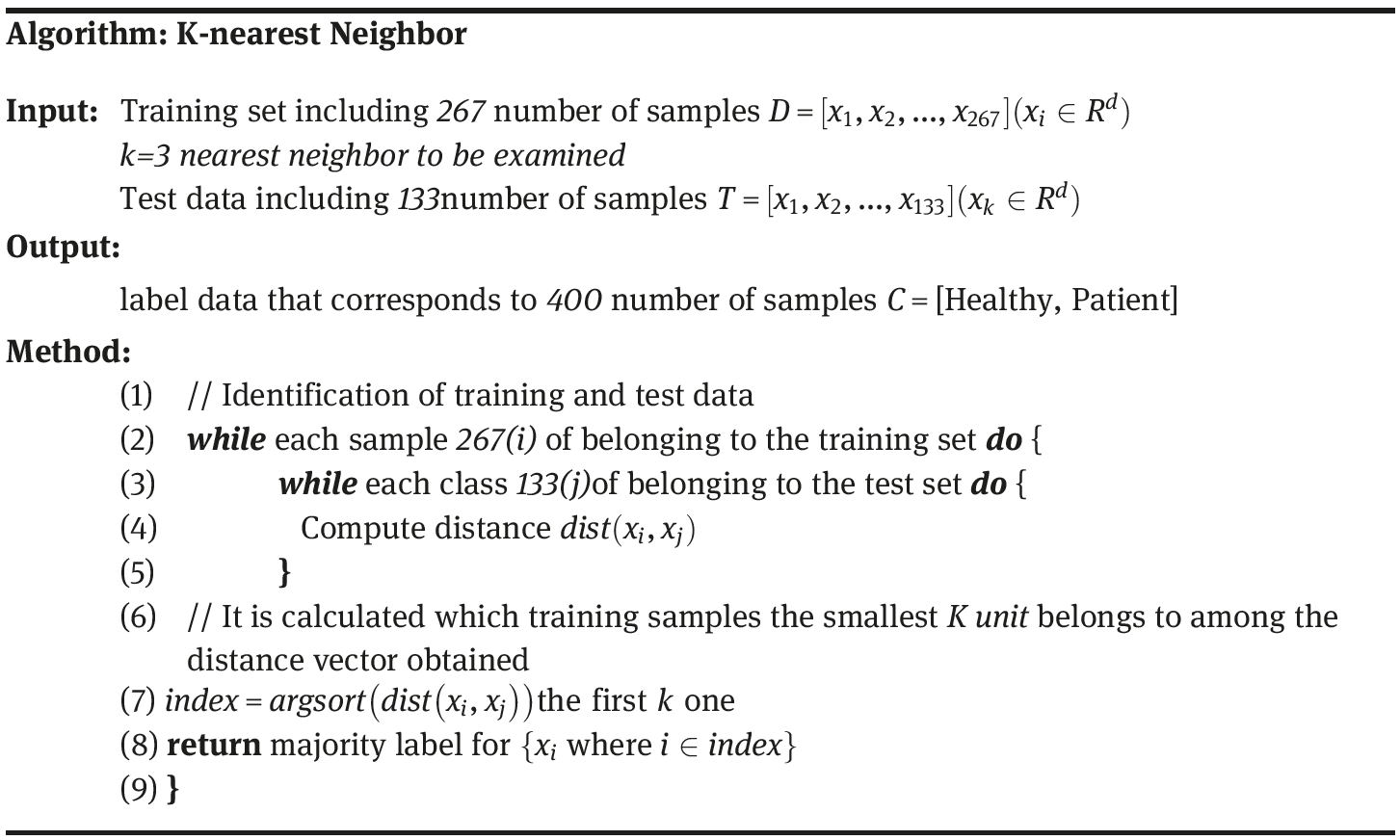
Step (2–3) Let us calculate the distance of the data to be classified to all the samples in accordance with the Euclid distance (eq. 9.1). D = (1, 1, ..., 1.4) and T = (3, 0, ..., 2.7), and let us calculate the distance between the data according to eq. (9.1).
Please bear in mind that the WAIS-R dataset comprised 21 attributes as administered to the individuals and we see that (1 ≤ i ≤ 21)
Step (5–9) Let us now find which class the smallest k number of neighbors belong to among the distance vector obtained dist(D, T) = [dist(D, T)1, dist(D, T)2, ..., dist(D, T)21]
How can we find the class of a test data belonging to WAIS-R test data according to k-nearest neighbor algorithm?
We can find the class label of a sample data belonging to WAIS-R dataset through the following steps specified below:
Step (1) The WAIS-R dataset samples are positioned to a 2D coordinate system as provided below: the blue dots represent Patient and the red ones represent Healthy class.
Step (2) Assume that the test data that we have classified on the coordinate plane where WAIS-R dataset exists corresponds to the green dot. The green dot selected from the dataset of WAIS-R dataset (Healthy, Patient) and data (whose class is not identified) has been obtained randomly from the dataset.

Step (3) Let us calculate the distances of the new test data to the training data according to Euclid distance formula (eq. 9.1). By ranking the distances calculated, find the two nearest neighbors to the test data.
Step (4) As the two nearest neighbors belong to the blue dot group, we can identify the class of our test data as blue that refers to Patient.

As a result, the data in the WAIS-R dataset with 33.33% portion allocated for the test procedure and classified as Patient and Healthy have been classified with an accuracy rate of 96.24% based on two nearest neighbor algorithm.
References
[1]Papadopoulos AN, Manolopoulos Y. Nearest neighbor search: A database perspective. USA: Springer Science & Business Media, 2006.
[2]Altman NS. An introduction to kernel and nearest-neighbor nonparametric regression. The American Statistician, 1992, 46(3), 175–185.
[3]Han J, Kamber M, Pei J, Data mining Concepts and Techniques. USA: The Morgan Kaufmann Series in Data Management Systems, Elsevier, 2012.
[4]Kramer O. Dimensionality reduction with unsupervised nearest neighbors. Berlin Heidelberg: Springer, 2013.
[5]Larose DT. Discovering knowledge in data: An introduction to data mining, USA: John Wiley & Sons, Inc., 90–106, 2005.
[6]Beyer K, Goldstein J, Ramakrishnan R, Shaft U. When is “nearest neighbor” meaningful?. In International conference on database theory. Springer: Berlin Heidelberg, 1999, January, 217–235.
[7]Simovici D, Djeraba C. Mathematical Tools for Data Mining. London: Springer, 2008.
[8]Khodabakhshi M, Fartash M. Fraud detection in banking using KNN (K-Nearest Neighbor) algorithm. In Intenational Conf. on Research in Science and Technology. London, United Kingdom, 2016.
[9]Wang SH, Zhang YD, Dong Z, Phillips P. Pathological Brain Detection. Singapore: Springer, 2018.
[10]Witten IH, Frank E, Hall MA, Pal CJ. Data Mining: Practical machine learning tools and techniques. USA: Morgan Kaufmann Series in Data Management Systems, Elsevier, 2016.
[11]Shmueli G, Bruce PC, Yahav I, Patel NR, Lichtendahl Jr, K. C. Data mining for business analytics: concepts, techniques, and applications in R. USA: John Wiley & Sons, Inc., 2017.
[12]Karaca Y, Cattani C, Moonis M, Bayrak Ş, Stroke Subtype Clustering by Multifractal Bayesian Denoising with Fuzzy C Means and K-Means Algorithms, Complexity,Hindawi, 2018.
[13]Rogers S, Girolami M. A first course in machine learning. USA: CRC Press, 2016.




