snow (“Simple Network of
Workstations”) is probably the most popular parallel programming package
available for R. It was written by Luke Tierney, A. J. Rossini, Na Li, and
H. Sevcikova, and is actively maintained by Luke Tierney. It is a mature
package, first released on the “Comprehensive R Archive Network” (CRAN) in
2003.
Motivation: You want to use a Linux cluster to run an R script faster. For example, you’re running a Monte Carlo simulation on your laptop, but you’re sick of waiting many hours or days for it to finish.
Solution: Use snow to run your R code on your company or
university’s Linux cluster.
Good because: snow fits well into a traditional cluster
environment, and is able to take advantage of high-speed communication
networks, such as InfiniBand, using MPI.
snow provides support for easily
executing R functions in parallel. Most of the parallel execution
functions in snow are variations of the
standard lapply() function, making
snow fairly easy to learn. To implement
these parallel operations, snow uses a
master/worker architecture, where the master sends tasks to the workers,
and the workers execute the tasks and return the results to the
master.
One important feature of snow is
that it can be used with different transport mechanisms to communicate
between the master and workers. This allows it to be portable, but still
take advantage of high-performance communication mechanisms if available.
snow can be used with socket
connections, MPI, PVM, or NetWorkSpaces. The socket transport doesn’t
require any additional packages, and is the most portable. MPI is
supported via the Rmpi package, PVM via
rpvm, and NetWorkSpaces via nws. The MPI transport is popular on Linux
clusters, and the socket transport is popular on multicore computers,
particularly Windows computers.[5]
snow is primarily intended to run
on traditional clusters and is particularly useful if MPI is available. It
is well suited to Monte Carlo simulations, bootstrapping, cross
validation, ensemble machine learning algorithms, and K-Means
clustering.
Good support is available for parallel random number generation,
using the rsprng and rlecuyer packages. This is very important when
performing simulations, bootstrapping, and machine learning, all of which
can depend on random number generation.
snow doesn’t provide mechanisms
for dealing with large data, such as distributing data files to the
workers. The input arguments must fit into memory when calling a snow function, and all of the task results are
kept in memory on the master until they are returned to the caller in a
list. Of course, snow can be used with
high-performance distributed file systems in order to operate on large
data files, but it’s up to the user to arrange that.
snow is available on CRAN, so it
is installed like any other CRAN package. It is pure R code and almost
never has installation problems. There are binary packages for both
Windows and Mac OS X.
Although there are various ways to install packages from CRAN, I
generally use the install.packages()
function:
install.packages("snow")It may ask you which CRAN mirror to use, and then it will download and install the package.
If you’re using an old version of R, you may get a message saying
that snow is not available. snow has required R 2.12.1 since version 0.3-5,
so you might need to download and install snow 0.3-3 from the CRAN package archives. In
your browser, search for “CRAN snow” and it will probably bring you to
snow’s download page on CRAN. Click on
the “snow archive” link, and then you can download snow_0.3-3.tar.gz. Or you can try directly
downloading it from:
http://cran.r-project.org/src/contrib/Archive/snow/snow_0.3-3.tar.gz
Once you’ve downloaded it, you can install it from the command line with:
% R CMD INSTALL snow_0.3-3.tar.gz
You may need to use the -l option
to specify a different installation directory if you don’t have permission
to install it in the default directory. For help on this command, use the
--help option. For more information on
installing R packages, see the section “Installing packages” in the “R
Installation and Administration” manual, written by the “R Development
Core Team”, and available from the R Project website.
Note
As a developer, I always use the most recent version of R. That
makes it easier to install packages from CRAN, since packages are only
built for the most recent version of R on CRAN. They keep around older
binary distributions of packages, but they don’t build new packages or
new versions of packages for anything but the current version of R. And
if a new version of a package depends on a newer version of R, as with
snow, you can’t even build it for
yourself on an older version of R. However, if you’re using R for
production use, you need to be much more cautious about upgrading to the
latest version of R.
To use snow with MPI, you will
also need to install the Rmpi package.
Unfortunately, installing Rmpi is a
frequent cause of problems because it has an external dependency on MPI.
For more information, see Installing Rmpi.
Fortunately, the socket transport can be used without installing any
additional packages. For that reason, I suggest that you start by using
the socket transport if you are new to snow.
Once you’ve installed snow, you
should verify that you can load it:
library(snow)
If that succeeds, you are ready to start using snow.
In order to execute any functions in parallel with snow, you must first create a
cluster object. The cluster object is used to
interact with the cluster workers, and is passed as the first argument
to many of the snow functions. You
can create different types of cluster objects, depending on the
transport mechanism that you wish to use.
The basic cluster creation function is makeCluster() which can create any type of
cluster. Let’s use it to create a cluster of four workers on the local
machine using the socket transport:
cl <- makeCluster(4, type="SOCK")
The first argument is the cluster specification, and the second is the cluster type. The interpretation of the cluster specification depends on the type, but all cluster types allow you to specify a worker count.
Socket clusters also allow you to specify the worker machines as a character vector. The following will launch four workers on remote machines:
spec <- c("n1", "n2", "n3", "n4")
cl <- makeCluster(spec, type="SOCK")The socket transport launches each of these workers via the
ssh command[6] unless the name is “localhost”, in which case makeCluster() starts the worker itself. For
remote execution, you should configure ssh to use password-less login. This can be
done using public-key authentication and SSH agents, which is covered in
chapter 6 of SSH, The
Secure Shell: The Definitive Guide (O’Reilly) and
many websites.
makeCluster() allows you to
specify addition arguments as configuration options. This is discussed
further in snow Configuration.
The type argument can be
“SOCK”, “MPI”, “PVM” or “NWS”. To create an MPI cluster with four
workers, execute:
cl <- makeCluster(4, type="MPI")
This will start four MPI workers on the local machine unless you make special provisions, as described in the section Executing snow Programs on a Cluster with Rmpi.
You can also use the functions
makeSOCKcluster(), makeMPIcluster(), makePVMcluster(), and makeNWScluster() to create specific types of
clusters. In fact, makeCluster() is
nothing more than a wrapper around these functions.
To shut down any type of cluster, use the stopCluster() function:
stopCluster(cl)
Some cluster types may be automatically stopped when the R session
exits, but it’s good practice to always call stopCluster() in snow scripts; otherwise, you risk leaking
cluster workers if the cluster type is changed, for example.
Note
Creating the cluster object can fail for a number of reasons, and is therefore a source of problems. See the section Troubleshooting snow Programs for help in solving these problems.
We’re finally ready to use snow
to do some parallel computing, so let’s look at a real example: parallel
K-Means. K-Means is a clustering algorithm that partitions rows of a
dataset into k clusters.[7] It’s an iterative algorithm, since it starts with a guess
of the location for each of the cluster centers, and gradually improves
the center locations until it converges on a solution.
R includes a function for performing K-Means clustering in the
stats package: the kmeans() function. One way of using the
kmeans() function is to specify the
number of cluster centers, and kmeans() will pick the starting points for the
centers by randomly selecting that number of rows from your dataset.
After it iterates to a solution, it computes a value called the
total within-cluster sum of squares. It then
selects another set of rows for the starting points, and repeats this
process in an attempt to find a solution with a smallest total
within-cluster sum of squares.
Let’s use kmeans() to generate
four clusters of the “Boston” dataset, using 100 random sets of
centers:
library(MASS) result <- kmeans(Boston, 4, nstart=100)
We’re going to take a simple approach to parallelizing kmeans() that can be used for parallelizing
many similar functions and doesn’t require changing the source code for
kmeans(). We simply call the kmeans() function on each of the workers using
a smaller value of the nstart
argument. Then we combine the results by picking the result with the
smallest total within-cluster sum of
squares.
But before we execute this in parallel, let’s try using this
technique using the lapply() function
to make sure it works. Once that is done, it will be fairly easy to
convert to one of the snow parallel
execution functions:
library(MASS) results <- lapply(rep(25, 4), function(nstart) kmeans(Boston, 4, nstart=nstart)) i <- sapply(results, function(result) result$tot.withinss) result <- results[[which.min(i)]]
We used a vector of four 25s to specify the nstart argument in order to get equivalent
results to using 100 in a single call to kmeans(). Generally, the length of this vector
should be equal to the number of workers in your cluster when running in
parallel.
Now let’s parallelize this algorithm. snow includes a number of functions that we
could use, including clusterApply(),
clusterApplyLB(), and parLapply(). For this example, we’ll use
clusterApply(). You call it exactly
the same as lapply(), except that it
takes a snow cluster object as the
first argument. We also need to load MASS on the workers, rather than on the
master, since it’s the workers that use the “Boston” dataset.
Assuming that snow is loaded
and that we have a cluster object named cl, here’s the parallel version:
ignore <- clusterEvalQ(cl, {library(MASS); NULL})
results <- clusterApply(cl, rep(25, 4), function(nstart) kmeans(Boston, 4,
nstart=nstart))
i <- sapply(results, function(result) result$tot.withinss)
result <- results[[which.min(i)]]clusterEvalQ() takes two
arguments: the cluster object, and an expression that is evaluated on
each of the workers. It returns the result from each of the workers in a
list, which we don’t use here. I use a compound expression to load
MASS and return NULL to avoid sending unnecessary data back to
the master process. That isn’t a serious issue in this case, but it can
be, so I often return NULL to be
safe.
As you can see, the snow
version isn’t that much different than the lapply() version. Most of the work was done in
converting it to use lapply().
Usually the biggest problem in converting from lapply() to one of the parallel operations is
handling the data properly and efficiently. In this case, the dataset
was in a package, so all we had to do was load the package on the
workers.
Note
The kmeans() function uses
the sample.int() function to choose
the starting cluster centers, which depend on the random number
generator. In order to get different solutions, the cluster workers
need to use different streams of random numbers. Since the workers are
randomly seeded when they first start generating random
numbers,[8] this example will work, but it is good practice to use a
parallel random number generator. See Random Number Generation for more
information.
In the last section we used the clusterEvalQ() function to initialize the
cluster workers by loading a package on each of them. clusterEvalQ() is very handy, especially for
interactive use, but it isn’t very general. It’s great for executing a
simple expression on the cluster workers, but it doesn’t allow you to
pass any kind of parameters to the expression, for example. Also,
although you can use it to execute a function, it won’t send that
function to the worker first,[9] as clusterApply()
does.
My favorite snow function for
initializing the cluster workers is clusterCall(). The arguments are pretty
simple: it takes a snow cluster
object, a worker function, and any number of arguments to pass to the
function. It simply calls the function with the specified arguments on
each of the cluster workers, and returns the results as a list. It’s
like clusterApply() without the
x argument, so it executes once for
each worker, like clusterEvalQ(),
rather than once for each element in x.
clusterCall() can do anything
that clusterEvalQ() does and
more.[10] For example, here’s how we could use clusterCall() to load the MASS package on the cluster workers:
clusterCall(cl, function() { library(MASS); NULL })This defines a simple function that loads the MASS package and returns NULL.[11] Returning NULL
guarantees that we don’t accidentally send unnecessary data transfer
back to the master.[12]
The following will load several packages specified by a character vector:
worker.init <- function(packages) {
for (p in packages) {
library(p, character.only=TRUE)
}
NULL
}
clusterCall(cl, worker.init, c('MASS', 'boot'))Setting the character.only
argument to TRUE makes library() interpret the argument as a
character variable. If we didn’t do that, library() would attempt to load a package
named p repeatedly.
Although it’s not as commonly used as clusterCall(), the clusterApply() function is also useful for
initializing the cluster workers since it can send different data to the
initialization function for each worker. The following creates a global
variable on each of the cluster workers that can be used as a unique
worker ID:
clusterApply(cl, seq(along=cl), function(id) WORKER.ID <<- id)
We introduced the clusterApply() function in the parallel
K-Means example. The next parallel execution function that I’ll discuss
is clusterApplyLB(). It’s very
similar to clusterApply(), but
instead of scheduling tasks in a round-robin
fashion, it sends new tasks to the cluster workers as they complete
their previous task. By round-robin, I mean that clusterApply() distributes the elements of
x to the cluster workers one at
a time, in the same way that cards
are dealt to players in a card game. In a sense, clusterApply() (politely)
pushes tasks to the workers, while clusterApplyLB() lets the workers pull tasks
as needed. That can be more efficient if some tasks take longer than
others, or if some cluster workers are slower.
To demonstrate clusterApplyLB(), we’ll execute Sys.sleep() on the workers, giving us complete control over the task lengths. Since our
real interest in using clusterApplyLB() is to improve performance,
we’ll use snow.time() to gather
timing information about the overall execution.[13] We will also use snow.time()’s plotting capability to visualize
the task execution on the workers:
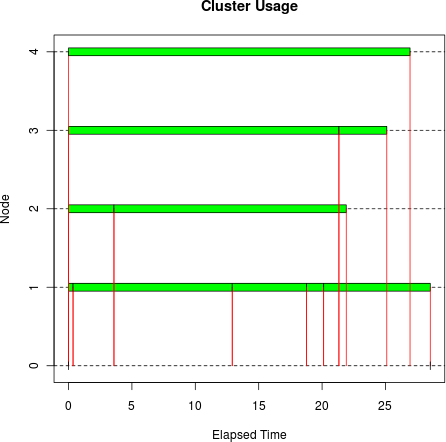
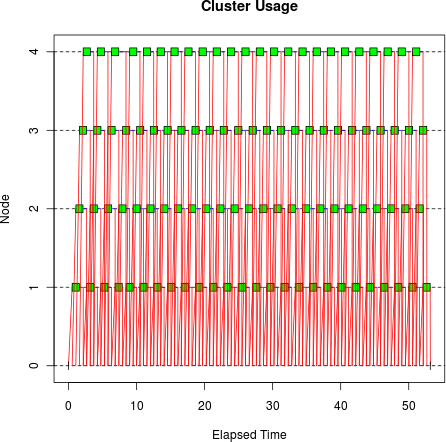
set.seed(7777442) sleeptime <- abs(rnorm(10, 10, 10)) tm <- snow.time(clusterApplyLB(cl, sleeptime, Sys.sleep)) plot(tm)
Ideally there would be solid horizontal bars for nodes 1 through 4
in the plot, indicating that the cluster workers were always busy, and
therefore running efficiently. clusterApplyLB() did pretty well, although
there was some wasted time at the end.
Now let’s try the same problem with clusterApply():[14]
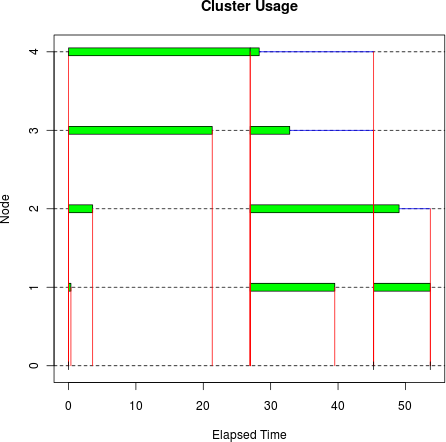
set.seed(7777442) sleeptime <- abs(rnorm(10, 10, 10)) tm <- snow.time(clusterApply(cl, sleeptime, Sys.sleep)) plot(tm)
As you can see, clusterApply()
is much less efficient than clusterApplyLB() in this example: it took 53.7
seconds, versus 28.5 seconds for clusterApplyLB(). The plot shows how much time
was wasted due to the round-robin scheduling.
But don’t give up on clusterApply(): it has its uses. It worked
fine in the parallel K-Means example because we had the same number of
tasks as workers. It is also used to implement the very useful parLapply() function, which we will discuss
next.[15]
Now that we’ve discussed and compared clusterApply() and clusterApplyLB(), let’s consider parLapply(), a third parallel lapply() function that has the same arguments
and basic behavior as clusterApply()
and clusterApplyLB(). But there is an
important difference that makes it perhaps the most generally useful of
the three.
parLapply() is a
high-level snow
function, that is actually a deceptively simple function wrapping an
invocation of clusterApply():
> parLapply function (cl, x, fun, ...) docall(c, clusterApply(cl, splitList(x, length(cl)), lapply, fun, ...)) <environment: namespace:snow>
Basically, parLapply() splits
up x into a list of subvectors, and
processes those subvectors on the cluster workers using lapply(). In effect, it is
prescheduling the work by dividing the tasks into
as many chunks as there are workers in the cluster. This is functionally
equivalent to using clusterApply()
directly, but it can be much more efficient, since there are fewer I/O
operations between the master and the workers. If the length of x is already equal to the number of workers,
then parLapply() has no advantage.
But if you’re parallelizing an R script that already uses lapply(), the length of x is often very large, and at any rate is
completely unrelated to the number of workers in your cluster. In that
case, parLapply() is a better
parallel version of lapply() than
clusterApply().
One way to think about it is that parLapply() interprets the x argument differently than clusterApply(). clusterApply() is
low-level, and treats x as a specification of the tasks to execute
on the cluster workers using fun.
parLapply() treats x as a source of disjoint input arguments to execute on the cluster workers
using lapply() and fun. clusterApply() gives you more control over what
gets sent to who, while parLapply()
provides a convenient way to efficiently divide the work among the
cluster workers.
An interesting consequence of parLapply()’s work scheduling is that it is
much more efficient than clusterApply() if you have many more tasks
than workers, and one or more large, additional arguments to pass to
parLapply(). In that case, the
additional arguments are sent to each worker only once, rather than
possibly many times. Let’s try doing that, using a slightly altered
parallel sleep function that takes a matrix as an argument:
bigsleep <- function(sleeptime, mat) Sys.sleep(sleeptime) bigmatrix <- matrix(0, 2000, 2000) sleeptime <- rep(1, 100)
I defined the sleeptimes to be small, many, and
equally sized. This will accentuate the performance differences between
clusterApply() and parLapply():
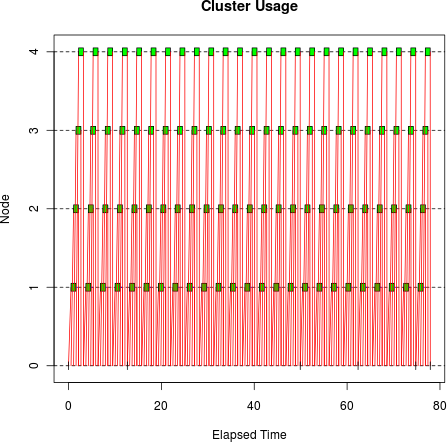
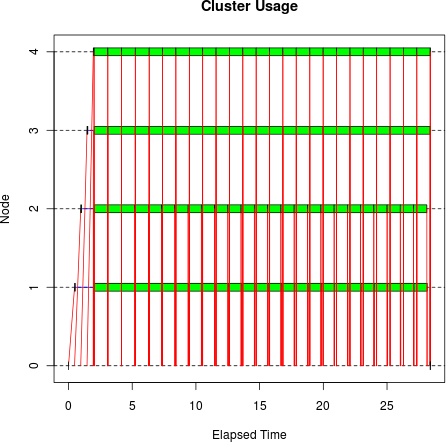
tm <- snow.time(clusterApply(cl, sleeptime, bigsleep, bigmatrix)) plot(tm)
This doesn’t look very efficient: you can see that there are many sends and receives between the master and the workers, resulting in relatively big gaps between the compute operations on the cluster workers. The gaps aren’t due to load imbalance as we saw before: they’re due to I/O time. We’re now spending a significant fraction of the elapsed time sending data to the workers, so instead of the ideal elapsed time of 25 seconds,[16] it’s taking 77.9 seconds.
Now let’s do the same thing using parLapply():
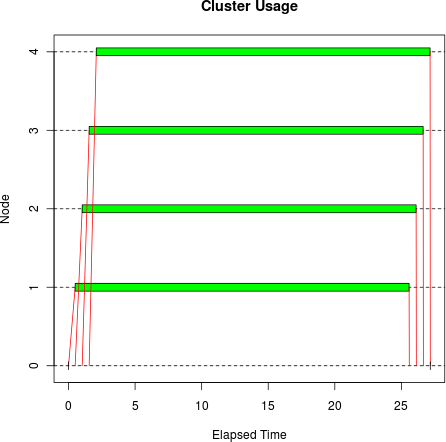
tm <- snow.time(parLapply(cl, sleeptime, bigsleep, bigmatrix)) plot(tm)
The difference is dramatic, both visually and in elapsed time: it
took only 27.2 seconds, beating clusterApply() by 50.7 seconds.
Keep in mind that this particular use of clusterApply() is bad: it is needlessly
sending the matrix to the worker with every task. There are various ways
to fix that, and using parLapply()
happens to work well in this case. On the other hand, if you’re sending
huge objects in x, then there’s not
much you can do, and parLapply()
isn’t going to help. My point is that parLapply() schedules work in a useful and
efficient way, making it probably the single most useful parallel
execution function in snow. When in
doubt, use parLapply().
In the previous section I showed you how parLapply() uses clusterApply() to implement a parallel
operation that solves a certain class of parallel program quite nicely.
Recall that parLapply() executes a
user-supplied function for each element of x just like clusterApply(). But what if we want the
function to operate on subvectors of x? That’s similar to what parLapply() does, but is a bit easier to
implement, since it doesn’t need to use lapply() to call the user’s function.
We could use the splitList()
function, like parLapply() does, but
that is a snow internal function.
Instead, we’ll use the clusterSplit()
function which is very similar, and slightly more convenient. Let’s try
splitting the sequence from 1 to 30 for our cluster using clusterSplit():
> clusterSplit(cl, 1:30) [[1]] [1] 1 2 3 4 5 6 7 8 [[2]] [1] 9 10 11 12 13 14 15 [[3]] [1] 16 17 18 19 20 21 22 [[4]] [1] 23 24 25 26 27 28 29 30
Since our cluster has four workers, it splits the sequence into a list of four nearly equal length vectors, which is just what we need.
Now let’s define parVapply() to
split x using clusterSplit(), execute the user function on
each of the pieces using clusterApply(), and combine the results using
do.call() and c():
parVapply <- function(cl, x, fun, ...) {
do.call("c", clusterApply(cl, clusterSplit(cl, x), fun, ...))
}Like parLapply(), parVapply() always issues the same number of
tasks as workers. But unlike parLapply(), the user-supplied function is
only executed once per worker. Let’s use parVapply() to compute the cube root of
numbers from 1 to 10 using the ^
function:
> parVapply(cl, 1:10, "^", 1/3) [1] 1.000000 1.259921 1.442250 1.587401 1.709976 1.817121 1.912931 2.000000 [9] 2.080084 2.154435
This works because the ^
function takes a vector as its first argument and returns a vector of
the same length.[17]
Note
This technique can be a useful for executing vector functions in
parallel. It may also be more efficient than using parLapply(), for example, but for any
function worth executing in parallel, the difference in efficiency is
likely to be small. And remember that most, if not all, vector
functions execute so quickly that it is never worth it to execute them
in parallel with snow. Such
fine-grained problems fall much more into the domain of multithreaded
computing.
We’ve talked about the advantages of parLapply() over clusterApply() at some length. In particular,
when there are many more tasks than cluster workers and the task objects
sent to the workers are large, there can be serious performance problems
with clusterApply() that are solved
by parLapply(). But what if the task
execution has significant variation so that we need load balancing?
clusterApplyLB() does load balancing,
but would have the same performance problems as clusterApply(). We would like a load balancing
equivalent to parLapply(), but there
isn’t one—so let’s write it.[18]
In order to achieve dynamic load balancing, it helps to have a
number of tasks that is at least a small integer multiple of the number
of workers. That way, a long task assigned to one worker can be offset
by many shorter tasks being done by other workers. If that is not the
case, then the other workers will sit idle while the one worker
completes the long task. parLapply()
creates exactly one task per worker, which is not what we want in this
case. Instead, we’ll first send the function and the fixed arguments to
the cluster workers using clusterCall(), which saves them in the global
environment, and then send the varying argument values using clusterApplyLB(), specifying a function that
will execute the user-supplied function along with the full collection
of arguments.
Here are the function definitions for parLapplyLB() and the two functions that it
executes on the cluster workers:
parLapplyLB <- function(cl, x, fun, ...) {
clusterCall(cl, LB.init, fun, ...)
r <- clusterApplyLB(cl, x, LB.worker)
clusterEvalQ(cl, rm('.LB.fun', '.LB.args', pos=globalenv()))
r
}
LB.init <- function(fun, ...) {
assign('.LB.fun', fun, pos=globalenv())
assign('.LB.args', list(...), pos=globalenv())
NULL
}
LB.worker <- function(x) {
do.call('.LB.fun', c(list(x), .LB.args))
}parLapplyLB() initializes the
workers using clusterCall(), executes
the tasks with clusterApplyLB(), cleans up the global
environment of the cluster workers with clusterEvalQ(), and finally returns the
task results.
That’s all there is to implementing a simple and efficient load
balancing parallel execution function. Let’s compare clusterApplyLB() to parLapplyLB() using the same test function
that we used to compare clusterApply() and parLapply(), starting with clusterApplyLB():
bigsleep <- function(sleeptime, mat) Sys.sleep(sleeptime) bigmatrix <- matrix(0, 2000, 2000) sleeptime <- rep(1, 100) tm <- snow.time(clusterApplyLB(cl, sleeptime, bigsleep, bigmatrix)) plot(tm)
There are lots of gaps in the execution bars due to high I/O time: the master is barely able to supply the workers with tasks. Obviously this problem isn’t going to scale to many more workers.
Now let’s try our new parLapplyLB() function:
tm <- snow.time(parLapplyLB(cl, sleeptime, bigsleep, bigmatrix)) plot(tm)
That took only 28.4 seconds versus 53.2 seconds for clusterApplyLB().
Notice that the first task on each worker has a short execution
time, but a long task send time, as seen by the
slope of the first four lines between the master (node 0) and the
workers (nodes 1-4). Those are the worker initialization tasks executed
by clusterCall() that send the large
matrix to the workers. The tasks executed via clusterApplyLB() were more efficient, as seen by
the vertical communication lines and the solid horizontal bars.
Note
By using short tasks, I was able to demonstrate a pretty noticeable difference in performance, but with longer tasks, the difference becomes less significant. In other words, we can realize decent efficiency whenever the time to compute a task significantly exceeds the time needed to send the inputs to and return the outputs from the worker evaluating the task.
Note
This section discusses a number of rather subtle points. An
understanding of these is not essential for basic snow use, but could be invaluable when
trying to debug more complicated usage scenarios. The reader may want
to skim through this on a first reading, but remember to return to it
if a seemingly obscure problem crops up.
Most of the parallel execution functions in snow take a function object as an argument,
which I call the worker function, since it is sent
to the cluster workers, and subsequently executed by them. In order to
send it to the workers, the worker function must be serialized into a
stream of bytes using the serialize()
function.[19] That stream of bytes is converted into a copy of the
original object using the unserialize() function.
In addition to a list of formal arguments and a body, the worker function includes a pointer to the environment in which it was created. This environment becomes the parent of the evaluation environment when the worker function is executed, giving the worker function access to non-local variables. Obviously, this environment must be serialized along with the rest of the worker function in order for the function to work properly after being unserialized.
However, environments are serialized in a special way in R. In general, the contents are included when an environment is serialized, but not always. Name space environments are serialized by name, not by value. That is, the name of the package is written to the resulting stream of bytes, not the symbols and objects contained in the environment. When a name space is unserialized, it is reconstructed by finding and loading the corresponding package. If the package cannot be loaded, then the stream of bytes cannot be unserialized. The global environment is also serialized by name, and when it is unserialized, the resulting object is simply a reference to the existing, unmodified global environment.
So what does this mean to you as a snow programmer? Basically, you must ensure
that all the variables needed to execute the worker function are
available after it has been unserialized on the cluster workers. If the
worker function’s environment is the global environment and the worker
function needs to access any variables in it, you need to send those
variables to the workers explicitly. This can be done, for example, by
using the clusterExport() function.
But if the worker function was created by another function, its
environment is the evaluation environment of the creator function when
the worker function was created. All the variables in this environment
will be serialized along with the worker function, and accessible to it
when it is executed by the cluster workers. This can be a handy way of
making variables available to the worker function, but if you’re not
careful, you could accidentally serialize large, unneeded objects along
with the worker function, causing performance to suffer. Also, if you
want the worker function to use any of the creator function’s arguments,
you need to evaluate those arguments before calling parLapply() or clusterApplyLB(); otherwise, you may not be
able to evaluate them successfully on the workers due to R’s lazy
argument evaluation.
Let’s look at a few examples to illustrate some of these issues.
We’ll start with a script that multiplies a vector x by a sequence of numbers:
a <- 1:4 x <- rnorm(4) clusterExport(cl, "x") mult <- function(s) s * x parLapply(cl, a, mult)
In this script, the function mult() is defined at the top level, so its
environment is the global environment.[20] Thus, x isn’t
serialized along with mult(), so we
need to send it to the cluster workers using the clusterExport() function. Of course, a more
natural solution in this case would be to include x as an explicit argument to mult(), and then parLapply() would send it to the workers for
us. However, using clusterExport()
could be more efficient if we were going to reuse x by calling mult() many times with parLapply().
Now let’s turn part of this script into a function. Although this
change may seem trivial, it actually changes the way mult() is serialized in parLapply():
pmult <- function(cl) {
a <- 1:4
x <- rnorm(4)
mult <- function(s) s * x
parLapply(cl, a, mult)
}
pmult(cl)Since mult() is created by
pmult(), all of pmult()’s local variables will be accessible
when mult() is executed by the
cluster workers, including x. Thus,
we no longer call clusterExport().
Pmult() would be more useful if
the values to be multiplied weren’t hardcoded, so let’s improve it by
passing a and x in as arguments:
pmult <- function(cl, a, x) {
x # force x
mult <- function(s) s * x
parLapply(cl, a, mult)
}
scalars <- 1:4
dat <- rnorm(4)
pmult(cl, scalars, dat)At this point, you may be wondering why x is on a line by itself with the cryptic
comment “force x”. Although it may look like it does nothing, this
operation forces x to be evaluated by
looking up the value of the variable dat (the actual argument corresponding to
x that is passed to the function when
pmult() is invoked) in the caller’s
execution environment. R uses lazy argument evaluation, and since
x is now an argument, we have to
force its evaluation before calling parLapply(); otherwise, the workers will
report that dat wasn’t found, since
they don’t have access to the environment where dat is defined. Note that they wouldn’t say
x wasn’t found: they would find
x, but wouldn’t be able to evaluate
it because they don’t have access to dat. By evaluating x before calling parLapply(), mult()’s environment will be serialized with
x set to the value of dat, rather than the symbol dat.
Notice in this last example that, in addition to x, a and
cl are also serialized along with
mult(). mult() doesn’t need to access them, but since
they are defined in pmult’s evaluation environment,
they will be serialized along with mult(). To prevent that, we can reset the environment of mult() to the global environment and pass
x to mult() explicitly:
pmult <- function(cl, a, x) {
mult <- function(s, x) s * x
environment(mult) <- .GlobalEnv
parLapply(cl, a, mult, x)
}
scalars <- 1:4
dat <- rnorm(4)
pmult(cl, scalars, dat)Of course, another way to achieve the same result is to create
mult() at the top level of the script
so that mult() is associated with the
global environment in the first place.
Unfortunately, you run into some tricky issues when sending function objects over the network. You may conclude that you don’t want to use the worker function’s environment to send data to your cluster workers, and that’s a perfectly reasonable position. But hopefully you now understand the issues well enough to figure out what methods work best for you.
As I mentioned previously, snow
is very useful for performing Monte Carlo simulations, bootstrapping,
and other operations that depend on the use of random numbers. When
running such operations in parallel, it’s important that the cluster
workers generate different random numbers; otherwise, the workers may
all replicate each other’s results, defeating the purpose of executing
in parallel. Rather than using ad-hoc schemes for seeding the workers
differently, it is better to use a parallel random number generator
package. snow provides support for
the rlecuyer and rsprng packages, both of which are available
on CRAN. With one of these packages installed on all the nodes of your
cluster, you can configure your cluster workers to use it via the
clusterSetupRNG() function. The
type argument specifies which
generator to use. To use rlecuyer,
set type to
RNGstream:
clusterSetupRNG(cl, type='RNGstream')
To use rsprng, set type to SPRNG:
clusterSetupRNG(cl, type='SPRNG')
You can specify a seed using the seed argument. rsprng uses a single integer for the seed,
while rlecuyer uses a vector of six
integers:
clusterSetupRNG(cl, type='RNGstream', seed=c(1,22,333,444,55,6))
Note
When using rsprng, a random
seed is used by default, but not with rlecuyer. If you want to use a random seed
with rlecuyer, you’ll have to
specify it explicitly using the seed argument.
Now the standard random number functions will use the specified parallel random number generator:
> unlist(clusterEvalQ(cl, rnorm(1))) [1] -1.0452398 -0.3579839 -0.5549331 0.7823642
If you reinitialize the cluster workers using the same seed, you will get the same random number from each of the workers.
We can also get reproducible results using clusterApply(), but not with clusterApplyLB() because clusterApply() always uses the same task
scheduling, while clusterApplyLB()
does not.[21]
snow includes a number of
configuration options for controlling the way the cluster is created.
These options can be specified as named arguments to the cluster
creation function (makeCluster(),
makeSOCKcluster(), makeMPIcluster(), etc.). For example, here is
the way to specify an alternate hostname for the master:
cl <- makeCluster(3, type="SOCK", master="192.168.1.100")
Note
The default value of master
is computed as Sys.info()[['nodename']]. However, there’s
no guarantee that the workers will all be able to resolve that name to
an IP address. By setting master to
an appropriate dot-separated IP address, you can often avoid hostname
resolution problems.
You can also use the setDefaultClusterOptions() function to change
a default configuration option during an R session. By default, the
outfile option is set to /dev/null, which causes all worker output to
be redirected to the null device (the proverbial bit bucket). To prevent
output from being redirected, you can change the default value of
outfile to the empty string:
setDefaultClusterOptions(outfile="")
This is a useful debugging technique which we will discuss more in Troubleshooting snow Programs.
Here is a summary of all of the snow configuration options:
Table 2-1. snow configuration options
| Name | Type | Description | Default value |
|---|---|---|---|
port | Integer | Port that the master listens on | 10187 |
timeout | Integer | Socket timeout in seconds | 31536000 (one year in seconds) |
master | String | Master’s hostname that workers connect to | Sys.info()["nodename"] |
homogeneous | Logical | Are workers homogeneous? | TRUE if R_SNOW_LIB set, else FALSE |
type | String | Type of cluster makeCluster should create | NULL, which is handled specially |
outfile | String | Worker log file | “/dev/null” “nul:” on Windows |
rhome | String | Home of R installation, used to locate R executable | $R_HOME |
user | String | User for remote execution | Sys.info()["user"] |
rshcmd | String | Remote execution command | “ssh” |
rlibs | String | Location of R packages | $R_LIBS |
scriptdir | String | Location of snow worker scripts | snow installation directory |
rprog | String | Path of R executable | $R_HOME/bin/R |
snowlib | String | Path of “library” where snow is installed | directory in which snow is installed |
rscript | String | Path of Rscript command | $R_HOME/bin/Rscript $R_HOME/bin/Rscript.exe on Windows |
useRscript | Logical | Should workers be started using Rscript command? | TRUE if file specified by Rscript exists |
manual | Logical | Should workers be started manually? | FALSE |
It is possible, although a bit tricky, to configure different
workers differently. I’ve done this when running a snow program in parallel on an ad-hoc
collection of workstations. In fact, there are two mechanisms available
for that with the socket transport. The first approach works for all the
transports. You set the homogeneous
option to FALSE, which causes
snow to use a special startup script
to launch the workers. This alternate script doesn’t assume that the
worker nodes are set up the same as the master node, but can look for
R or Rscript in the user’s PATH, for example. It also supports the use of
environment variables to configure the workers, such as R_SNOW_RSCRIPT_CMD and R_SNOW_LIB to specify the path of the Rscript command and the snow installation directory. These environment
variables can be set to appropriate values in the user’s environment on
each worker machine using the shell’s start up scripts.
The second approach to heterogeneous configuration only works with
the socket and nws transports. When
you call makeSOCKcluster(), you
specify the worker machines as a list of lists. In this case, the
hostname of the worker is specified by the host element of each sublist. The other
elements of the sublists are used to override the corresponding option
for that worker.
Let’s say we want to create a cluster with two workers: n1 and n2, but we need to log in as a different user on machine n2:
> workerList <- list(list(host = "n1"), list(host = "n2", user = "steve")) > cl <- makeSOCKcluster(workerList) > clusterEvalQ(cl, Sys.info()[["user"]]) [[1]] [1] "weston" [[2]] [1] "steve" > stopCluster(cl)
It can also be useful to set the outfile option differently to avoid file
conflicts between workers:
> workerList <- list(list(host = "n1", outfile = "n1.log", user = "weston"),
+ list(host = "n2", outfile = "n2-1.log"),
+ list(host = "n2", outfile = "n2-2.log"))
> cl <- makeSOCKcluster(workerList, user = "steve")
> clusterEvalQ(cl, Sys.glob("*.log"))
[[1]]
[1] "n1.log"
[[2]]
[1] "n2-1.log" "n2-2.log"
[[3]]
[1] "n2-1.log" "n2-2.log"
> stopCluster(cl)This also demonstrates that different methods for setting options can be used together. The machine-specific option values always take precedence.
Note
I prefer to use my ssh config
file to specify a different user for different hosts, but obviously
that doesn’t help with setting outfile.
As I mentioned previously, installing Rmpi can be problematic because it depends on
MPI being previously installed. Also, there are multiple MPI
distributions, and some of the older distributions have compatibility
problems with Rmpi. In general, Open
MPI is the preferred MPI distribution. Fortunately, Open MPI is readily
available for modern Linux systems. The website for the Open MPI Project
is http://www.open-mpi.org/.
Another problem is that there isn’t a binary distribution of
Rmpi available for Windows. Thus,
even if you have MPI installed on a Windows machine, you will also need
to install Rmpi from the source
distribution, which requires additional tools that may also need to be
installed. For more information on installing Rmpi on Windows, see the documentation in the
Rmpi package. That’s beyond the scope
of this book.
Installation of Rmpi on the Mac
was quite simple on Mac OS X 10.5 and 10.6, both of which came with Open
MPI, but unfortunately, Apple stopped distributing it in Mac OS X 10.7.
If you’re using 10.5 or 10.6, you can (hopefully) install Rmpi quite easily:[22]
install.packages("Rmpi")If you’re using Mac OS X 10.7, you’ll have to install Open MPI
first, and then you’ll probably have to build Rmpi from the source distribution since the
binary distribution probably won’t be compatible with your installation
of Open MPI. I’ll discuss installing Rmpi from the source distribution shortly, but
not Open MPI.
On Debian/Ubuntu, Rmpi is
available in the “r-cran-rmpi” Debian package, and can be installed with
apt-get. That’s the most foolproof
way to install Rmpi on Ubuntu, for
example, since apt-get will
automatically install a compatible version of MPI, if necessary.
For non-Debian based systems, I recommend that you install Open
MPI with your local packaging tool, and then try to use install.packages() to install Rmpi. This will fail if the configuration
script can’t find the MPI installation. In that case you will have to
download the source distribution, and install it using a command such
as:
% R CMD INSTALL --configure-args="--with-mpi=$MPI_PATH" Rmpi_0.5-9.tar.gz
where the value of MPI_PATH is
the directory containing the Open MPI lib and include directories.[23] Notice that this example uses the --configure-args argument to pass the --with-mpi argument to Rmpi’s configure script. Another important
configure argument is --with-Rmpi-type, which may need to be set to
“OPENMPI”, for example.
As I’ve said, installing Rmpi
from source can be difficult. If you run into problems and don’t want to
switch to Debian/Ubuntu, your best bet is to post a question on the R
project’s “R-sig-hpc” mailing list. You can find it by clicking on the
“Mailing Lists” link on the R project’s home page.
Throughout this chapter I’ve been using the socket transport
because it doesn’t require any additional software to install, making it
the most portable snow transport.
However, the MPI transport is probably the most popular, at least on
clusters. Of course, most of what we’ve discussed is independent of the
transport. The difference is mostly in how the cluster object is created
and how the snow script is
executed.
To create an MPI cluster object, set the type argument of makeCluster() to MPI or use
the makeMPIcluster() function. If
you’re running interactively, you can create an MPI cluster object with
four workers as follows:
cl <- makeCluster(4, type="MPI")
This is equivalent to:
cl <- makeMPIcluster(4)
This creates a spawned cluster, since the
workers are all started by snow for
you via the mpi.comm.spawn()
function.
Notice that we don’t specify which machines to use, only the
number of workers. For that reason, I like to compute the worker count
using the mpi.universe.size()
function, which returns the size of the initial runtime
environment.[24] Since the master process is included in that size, the
worker count would be computed as mpi.universe.size() - 1.[25]
We shut down an MPI cluster the same as any cluster:
stopCluster(cl)
As you can see, there isn’t much to creating an MPI cluster
object. You can specify configuration options, just as with a socket
cluster, but basically it is very simple. However, you should be aware
that the cluster workers are launched differently depending on how the R
script was executed. If you’re running interactively, for example, the
workers will always be started on the local machine. The only way that I
know of to start the workers on remote machines is to execute the R
interpreter using a command such as mpirun, mpiexec, or in the case of Open MPI, orterun.
As I noted previously, you can’t specify the machines on which to
execute the workers with makeMPIcluster(). That is done with a separate
program that comes with your MPI distribution. Open MPI comes with three
utilities for executing MPI programs: orterun, mpirun, and mpiexec, but they all work in exactly the same
way,[26] so I will refer to orterun for the rest of this
discussion.
orterun doesn’t know anything
about R or R scripts, so we need to use orterun to execute the R interpreter, which in
turn executes the R script. Let’s start by creating an R script (Example 2-1), which I’ll call mpi.R.
Example 2-1. mpi.R
library(snow) library(Rmpi) cl <- makeMPIcluster(mpi.universe.size() - 1) r <- clusterEvalQ(cl, R.version.string) print(unlist(r)) stopCluster(cl) mpi.quit()
This is very similar to our very first example, except that it
loads the Rmpi package, calls
makeMPIcluster() rather than makeSOCKcluster(), and calls mpi.quit() at the end. Loading Rmpi isn’t strictly necessary, since calling
makeMPIcluster() will automatically
load Rmpi, but I like to do it
explicitly. makeMPIcluster() creates
the MPI cluster object, as discussed in the previous section. mpi.quit() terminates the MPI execution
environment, detaches the Rmpi
package, and quits R, so it should always go at the
end of your script. This is often left out, but I believe it is good
practice to call it.[27] I’ve gotten very stern warning messages from orterun in some cases when I failed to call
mpi.quit().
To execute mpi.R using the
local machine as the master, and n1, n2, n3 and n4 as the workers, we
can use the command:[28]
% orterun -H localhost,n1,n2,n3,n4 -n 1 R --slave -f mpi.R
The -H option specifies the
list of machines available for execution. By using -n 1, orterun will only execute the command R --slave -f mpi.R on the first machine in the
list, which is localhost in this example. This process is the master,
equivalent to the interactive R session in our previous snow examples. When the master executes
makeMPIcluster(mpi.universe.size() -
1), four workers will be spawned. orterun will execute these workers on machines
n1, n2, n3 and n4, since they are next in line to receive a
process.
Those are the basics, but there are a few other issues to bear in
mind. First, the master and the worker processes have their working
directory set to the working directory of the process executing orterun. That’s no problem for the master in
our example, since the master runs on the same machine as orterun. But if there isn’t a directory with
the same path on any of the worker machines, you will get an error. For
that reason, it is useful to work from a directory that is shared across
the cluster via a network file system. That isn’t necessary, however. If
you specify the full path to the R script, you could use the orterun -wdir option to set the working directory to
/tmp:
% orterun -wdir /tmp -H localhost,n1,n2,n3,n4 -n 1 R --slave -f ~/mpi.R
This example still assumes that R is in your search path on localhost. If it isn’t, you can specify the full path of the R interpreter on localhost.
That can solve some of the orterun related problems, but snow still makes a number of assumptions about
where to find things on the workers as well. See snow Configuration for more information.
Many cluster administrators require that all parallel programs be executed via a batch queueing system. There are different ways that this can be done, and different batch queueing systems, but I will describe a method that has been commonly used for a long time, and is supported by many batch queueing systems, such as PBS/TORQUE, SGE and LSF.
Basically you submit a shell script, and the shell script executes
your R script using orterun as we
described in the section Executing snow Programs on a Cluster with Rmpi. When you submit the
shell script, you tell the batch queueing system how many nodes you want
using the appropriate argument to the submit command. The shell script
may need to read an environment variable to learn what nodes it can
execute on, and then pass that information on to the orterun command via an argument such as
-hostfile or -H.
Of course the details vary depending on the batch queueing system, MPI distribution, and cluster configuration. As an example, I’ll describe how this can be done using PBS/TORQUE and Open MPI.
It’s actually very simple to use PBS/TORQUE with Open MPI, since
Open MPI automatically gets the list of hosts using the environment
variables set by PBS/TORQUE.[29] The code in Example 2-2 simplifies the
orterun command used in the
script.
Example 2-2. batchmpi.sh
#!/bin/sh #PBS -N SNOWMPI #PBS -j oe cd $PBS_O_WORKDIR orterun -n 1 /usr/bin/R --slave -f mpi.R > mpi-$PBS_JOBID.out 2>&1
This script uses PBS directives to specify the name of the job,
and to merge the job’s standard output and standard error. It then
cd’s to the directory from which you
submitted the job, which is helpful for finding the mpi.R script. Finally it uses orterun to execute mpi.R.
We submit batchmpi.sh using the
PBS/TORQUE qsub command:
% qsub -q devel -l nodes=2:ppn=4 batchmpi.sh
This submits the shell script to the devel queue, requesting two nodes with four
processors per node. The -l option is
used to specify the resources needed by the job. The resource
specifications vary from cluster to cluster, so talk to your cluster
administrator to find out how you should specify the number of nodes and
processors.
If you’re using LSF or SGE, you will probably need to specify the
hosts via the orterun -hostfile or -H option. For LSF, use the
bsub -n option to specify the number of cores, and
the LSB_HOSTS environment variable to
get the allocated hosts. With SGE, use the qsub -pe
option and the PE_HOSTFILE
environment variable. The details are different, but the basic idea is
the same.
Unfortunately, a lot of things can go wrong when using snow. That’s not really snow’s fault: there’s just a lot of things
that have to be set up properly, and if the different cluster nodes are
configured differently, snow may have
trouble launching the cluster workers. It’s possible to configure
snow to deal with heterogeneous
clusters.[30] Fortunately, if your cluster is already used for parallel
computing, there’s a good chance it is already set up in a clean,
consistent fashion, and you won’t run into any problems when using
snow.
Obviously you need to have R and snow installed on all of the machines that
you’re attempting to use for your cluster object. You also need to have
ssh servers running on all of the
cluster workers if using the socket transport, for instance.
There are several techniques available for finding out more information about what is going wrong.
When using the socket transport, the single most useful method of
troubleshooting is manual mode. In manual mode, you
start the workers yourself, rather than having snow start them for you. That allows you to
run snow jobs on a cluster that
doesn’t have ssh servers, for
example. But there are also a few other advantages to manual mode. For
one thing, it makes it easier to see error messages. Rather than
searching for them in log files, they can be displayed right in your
terminal session.
To enable manual mode, set the manual option to TRUE when creating the socket cluster object.
I also recommend specifying outfile="", which prevents output from being
redirected:
cl <- makeCluster(2, type="SOCK", manual=TRUE, outfile="")
makeCluster() will display the
command to start each of the workers. For each command, I open a new
terminal window, ssh to the specified
machine,[31] and cut and paste the specified command into the
shell.
In many cases, you’ll get an error message as soon as you execute
one of these commands, and the R session will exit. In that case, you
need to figure out what caused the error, and solve the problem. That
may not be simple, but at least you have something better to search for
than “makeCluster hangs.” But very often, the error is pretty obvious,
like R or snow isn’t installed. Also,
snow may not guess the right hostname
for the workers to use to connect back to the master process. In this
case, R starts up and snow runs, but
nothing happens. You can use your terminal window to use various network
tools (nslookup, ping) to diagnose this problem.
Let’s create a socket cluster using manual mode and examine the output:
> cl <- makeCluster(c('n1', 'n2'), type="SOCK", manual=TRUE, outfile="")
Manually start worker on n1 with
/usr/lib/R/bin/Rscript /usr/lib/R/site-library/snow/RSOCKnode.R
MASTER=beard PORT=10187 OUT= SNOWLIB=/usr/lib/R/site-libraryThe argument MASTER=beard
indicates that the value of the master option is “beard.” You can now use the
ping command from your terminal
window on n1 to see if the master is
reachable from n1 by that name.
Here’s the kind of output that you should see:
n1% ping beard PING beard (192.168.1.109) 56(84) bytes of data. 64 bytes from beard (192.168.1.109): icmp_req=1 ttl=64 time=0.020 ms
This demonstrates that n1 is
able to resolve the name “beard,” knows a network route to that IP
address, can get past any firewall, and is able to get a reply from the
master machine.[32]
But if ping issues the error
message “ping: unknown host beard”, then you have a hostname resolution
problem. Setting the master option to
a different value when creating the cluster might fix the problem. Other
errors may indicate a networking problem that can be fixed by your
sysadmin.
If the value of master seems
good, you should execute the command displayed by makeCluster() in hopes of getting a useful
error message. Note that many of these problems could occur using any
snow transport, so running a simple
snow test code using the socket
transport and manual mode can be an effective means to ensure a good
setup even if you later intend to use a different transport.
The outfile option in itself is
also useful for troubleshooting. It allows you to redirect debug and
error messages to a specified file. By default, output is redirected to
/dev/null. I often use an empty
string ("") to prevent any
redirection, as we described previously.
Here are some additional troubleshooting tips:
Start by running on only one machine to make sure that works
Manually
sshto all of the workers from the master machineSet the
masteroption to a value that all workers can resolve, possibly using a dot-separated IP addressRun your job from a directory that is available on all machines
Check if there are any firewalls that might interfere
snow is a fairly high-level
package, since it doesn’t focus on low-level communication operations, but
on execution. It provides a useful variety of functions that support
embarrassingly parallel computation.
Communications difficulties:
snow doesn’t provide functions for
explicitly communicating between the master and workers, and in fact, the
workers never communicate between themselves. In order to communicate
between workers, you would have to use functions in the underlying
communication package. Of course, that would make your program less
portable, and more complicated. A package that needed to do that would
probably not use snow, but use a
package like nws or Rmpi directly.
In this chapter, you got a crash course on the snow package, including some advanced topics
such as running snow programs via a
batch queueing system. snow is a
powerful package, able to run on clusters with hundreds of nodes. But if
you’re more interested in running on a quad-core laptop than a
supercomputer, the next chapter on the multicore package will be of particular
interest to you.
[5] The multicore package is
generally preferred on multicore computers, but it isn’t supported on
Windows. See Chapter 3 for more information on the
multicore package.
[6] This can be overridden via the rshcmd option, but the specified command
must be command line-compatible with ssh.
[7] These clusters shouldn’t be confused with cluster objects and cluster workers.
[8] All R sessions are randomly seeded when they first generate
random numbers, unless they were restored from a previous R
session that generated random numbers. snow
workers never restore previously saved data, so they are always
randomly seeded.
[9] How exactly snow sends
functions to the workers is a bit complex, raising issues of
execution context and environment. See Functions and Environments for
more information.
[10] This is guaranteed since clusterEvalQ() is implemented using
clusterCall().
[11] Defining anonymous functions like this is very useful, but can be a source of performance problems due to R’s scoping rules and the way it serializes functions. See Functions and Environments for more information.
[12] The return value from library() isn’t big, but if the
initialization function was assigning a large matrix to a variable,
you could inadvertently send a lot of data back to the master,
significantly hurting the performance of your program.
[13] snow.time() is available in
snow as of version 0.3-5.
[14] I’m setting the RNG seed so we get the same value of
sleeptime as in the previous example.
[15] It’s also possible that the extra overhead in clusterApplyLB() to determine which worker
is ready for the next task could make clusterApply() more efficient in some
case, but I’m skeptical.
[16] The ideal elapsed time is sum(sleeptime) / length(cl).
[17] Normally the second argument to ^ can have the same length as the first,
but it must be length one in this example because parVapply() only splits the first
argument.
[18] A future release of snow
could optimize clusterApplyLB()
by not sending the function and constant arguments to the workers in
every task. At that point, this example will lose any practical
value that it may have.
[19] Actually, if you specify the worker function by name, rather
than by providing the definition of the function, most of the
parallel execution functions (parLapply() is currently an exception)
will use that name to look up that function in the worker processes,
thus avoiding function serialization.
[20] You can verify this with the command environment(mult).
[21] Actually, you can achieve reproducibility with clusterApplyLB() by setting the seed to a
task specific value. This can be done by adding the operation to the
beginning of the worker function, or if using a function from a
library, wrapping that function in a new function that sets the seed
and then calls the library function.
[22] It’s possible that newer versions of Rmpi won’t be built for the Mac on CRAN
because it won’t work on Mac OS X 10.7, but it’s still available as
I’m writing this in September 2011.
[23] I use the command locate
include/mpi.h to find this directory. On my machine, this
returns /usr/lib/openmpi/include/mpi.h, so I set
MPI_PATH to /usr/lib/openmpi.
[24] mpi.universe.size() had a
bug in older versions of Rmpi, so
you may need to upgrade to Rmpi 0.5-9.
[25] I don’t use mpi.universe.size() when creating an MPI
cluster in an interactive session, since in that context, mpi.universe.size() returns 1, which would
give an illegal worker count of zero.
[26] orterun, mpirun, and mpiexec are in fact the same program in
Open MPI.
[27] You can use mpi.finalize()
instead, which doesn’t quit R.
[28] The orterun command in Open
MPI accepts several different arguments to specify the host list and
the number of workers. It does this to be compatible with previous
MPI distributions, so don’t be confused if you’re used to different
argument names.
[29] Actually, it’s possible to configure Open MPI without support
for PBS/TORQUE, in which case you’ll have to include the arguments
-hostfile $PBS_NODEFILE when
executing orterun.
[30] We discuss heterogeneous configuration in snow Configuration.
[31] If ssh fails at this point,
you may have found your problem.
[32] Of course, just because ping can get past a firewall doesn’t mean
that snow can. As you can see
from the manual mode output, the master process is listening on port
10187, so you may have to configure your firewall to allow
connections on that port. You could try the command telnet beard 10187 as a further
test.