Chapter 9. Clustering
Up until this point we have been solving problems of fitting a function to a set of data. For instance, given previously observed mushroom attributes and edibleness, how would we classify new mushrooms? Or, given a neighborhood, what should the house value be?
This chapter talks about a completely different learning problem: clustering. This is a subset of unsupervised learning methods and is useful in practice for understanding data at its core.
If you’ve been to a library you’ve seen clustering at work. The Dewey Decimal system is a form of clustering. Dewey came up with a system that attached numbers of increasing granularity to categories of books, and it revolutionized libraries.
We will talk about what it means to be unsupervised and what power exists in that, as well as two clustering algorithms: K-Means and expectation maximization (EM) clustering. We will also address two other issues associated with clustering and unsupervised learning:
-
How do you test a clustering algorithm?
-
The impossibility theorem.
Studying Data Without Any Bias
If I were to give you an Excel spreadsheet full of data, and instead of giving you any idea as to what I’m looking for, just asked you to tell me something, what could you tell me? That is what unsupervised learning aims to do: study what the data is about.
A more formal definition would be to think of unsupervised learning as finding the best function f such that f(x) = x. At first glance, wouldn’t x = x? But it’s more than just that—you can always map data onto itself—but what unsupervised learning does is define a function that describes the data.
What does that mean?
Unsupervised learning is trying to find a function that generalizes the data to some degree. So instead of trying to fit it to some classification or number, instead we are just fitting the function to describe the data. This is essential to understand since it gives us a glimpse as to how to test this.
Let’s dive into an example.
User Cohorts
Grouping people into cohorts makes a lot of business and marketing sense. For instance, your first customer is different from your ten thousandth customer or your millionth customer. This problem of defining users into cohorts is a common one. If we were able to effectively split our customers into different buckets based on behavior and time of signup, then we could better serve them by diversifying our marketing strategy.
The problem is that we don’t have a predefined label for customer cohorts. To get over this problem you could look at what month and year they became a customer. But that is making a big assumption about that being the defining factor that splits customers into groups. What if time of first purchase had nothing to do with whether they were in one cohort or the other? For example, they could only have made one purchase or many.
Instead, what can we learn from the data? Take a look at Table 9-1. Let’s say we know when they signed up, how much they’ve spent, and what their favorite color is. Assume also that over the last two years we’ve only had 10 users sign up (well, I hope you have more than that over two years, but let’s keep this simple).
| User ID | Signup date | Money spent | Favorite color |
|---|---|---|---|
1 |
Jan 14 |
40 |
N/A |
2 |
Nov 3 |
50 |
Orange |
3 |
Jan 30 |
53 |
Green |
4 |
Oct 3 |
100 |
Magenta |
5 |
Feb 1 |
0 |
Cyan |
6 |
Dec 31 |
0 |
Purple |
7 |
Sep 3 |
0 |
Mauve |
8 |
Dec 31 |
0 |
Yellow |
9 |
Jan 13 |
14 |
Blue |
10 |
Jan 1 |
50 |
Beige |
Given these data, we want to learn a function that describes what we have. Looking at these rows, you notice that the favorite colors are irrelevant data. There is no information as to whether users should be in a cohort. That leaves us with Money spent and Signup date. There seems to be a group of users who spend money, and one of those who don’t. That is useful information. In the Signup date column you’ll notice that there are a lot of users who sign up around the beginning of the year and end of the previous one, as well as around September, October, or November.
Now we have a choice: whether we want to find the gist of this data in something compact or find a new mapping of this data onto a transformation. Remember the discussion of kernel tricks in Chapter 7? This is all we’re doing—mapping this data onto a new dimension. For the purposes of this chapter we will delve into a new mapping technique: in Chapter 10, on data extraction and improvement, we’ll delve into compaction of data.
Let’s imagine that we have 10 users in our database and have information on when they signed up, and how much money they spent. Our marketing team has assigned them manually to cohorts (Table 9-2).
| User ID | Signup date (days to Jan 1) | Money spent | Cohort |
|---|---|---|---|
1 |
Jan 14 (13) |
40 |
1 |
2 |
Nov 3 (59) |
50 |
2 |
3 |
Jan 30 (29) |
53 |
1 |
4 |
Oct 3 (90) |
100 |
2 |
5 |
Feb 1 (31) |
0 |
1 |
6 |
Dec 31 (1) |
0 |
1 |
7 |
Sep 3 (120) |
0 |
2 |
8 |
Dec 31 (1) |
0 |
1 |
9 |
Jan 13 (12) |
14 |
1 |
10 |
Jan 1 (0) |
50 |
1 |
We have divided the group into two groups where seven users are in group 1, which we could call the beginning-of-the-year signups, and end-of-the-year signups are in group 2.
But there’s something here that doesn’t sit well. We assigned the users to different clusters, but didn’t really test anything—what to do?
Testing Cluster Mappings
Testing unsupervised methods doesn’t have a good tool such as cross-validation, confusion matrices, ROC curves, or sensitivity analysis, but they still can be tested, using one of these two methods:
-
Determining some a priori fitness of the unsupervised learning method
-
Comparing results to some sort of ground truth
Fitness of a Cluster
Domain knowledge can become very useful in determining the fitness of an unsupervised model. For instance, if we want to find things that are similar, we might use some sort of distance-based metric. If instead we wish to determine independent factors of the data, we might calculate fitness based on correlation or covariance.
Possible fitness functions include:
Mean distances from centroid, or from all points in a cluster, are almost baked into algorithms that we will be talking about such as K-Means or EM clustering, but the silhouette coefficient is an interesting take on fitness of cluster mappings.
Silhouette Coefficient
The silhouette coefficient evaluates cluster performance without ground truth (i.e., data that has been provided through direct observation versus inferred observations) by looking at the relation of the average distance inside of a cluster versus the average distance to the nearest cluster (Figure 9-1).
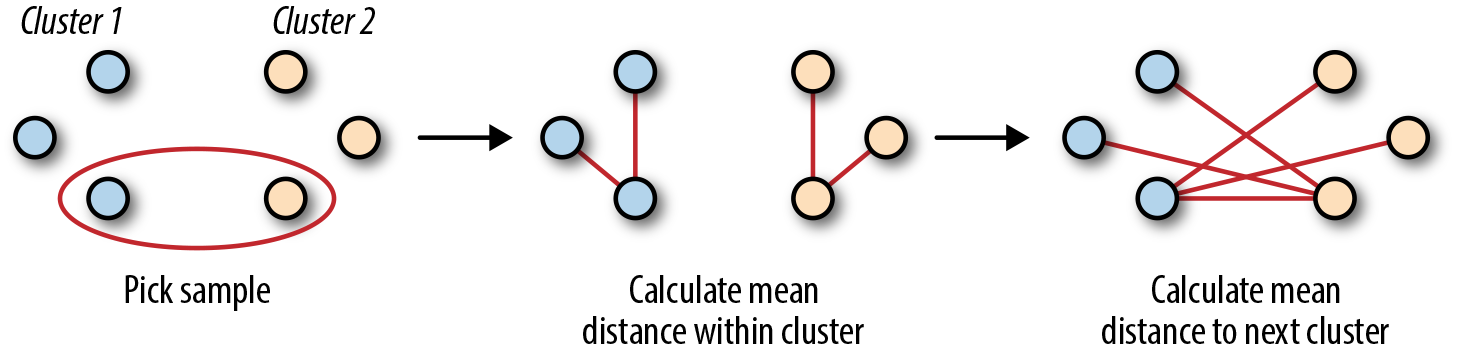
Figure 9-1. Silhouette coefficient visually
Mathematically the metric is:
where a is the average distance between a sample and all other points in that cluster and b is the same sample’s mean distance to the next nearest cluster points.
This coefficient ends up becoming useful because it will show fitness on a scale of –1 to 1 while not requiring ground truth.
Comparing Results to Ground Truth
In practice many times machine learning involves utilizing ground truth, which is something that we can usually find through trained data sets, humans, or other means like test equipment. This data is valuable in testing our intuition and determining how fitting our model is.
Clustering can be tested using ground truth using the following means:
-
Rand index
-
Mutual information
-
Homogeneity
-
Completeness
-
V-measure
-
Fowlkes-Mallows score
All of these methods can be extremely useful in determining how fit a model is. scikit-learn implements all of these and can easily be used to determine a score.
K-Means Clustering
There are a lot of clustering algorithms like linkage clustering or Diana, but one of the most common is K-Means clustering. Using a predefined K, which is the number of clusters that one wants to split the data into, K-Means will find the most optimal centroids of clusters. One nice property of K-Means clustering is that the clusters will be strict, spherical in nature, and converge to a solution.
In this section we will briefly talk about how K-Means clustering works.
The K-Means Algorithm
The K-Means algorithm starts with a base case. Pick K random points in the data set and define them as centroids. Next, assign each point to a cluster number that is closest to each different centroid. Now we have a clustering based on the original randomized centroid. This is not exactly what we want to end with, so we update where the centroids are using a mean of the data. At that point we repeat until the centroids no longer move (Figure 9-2).
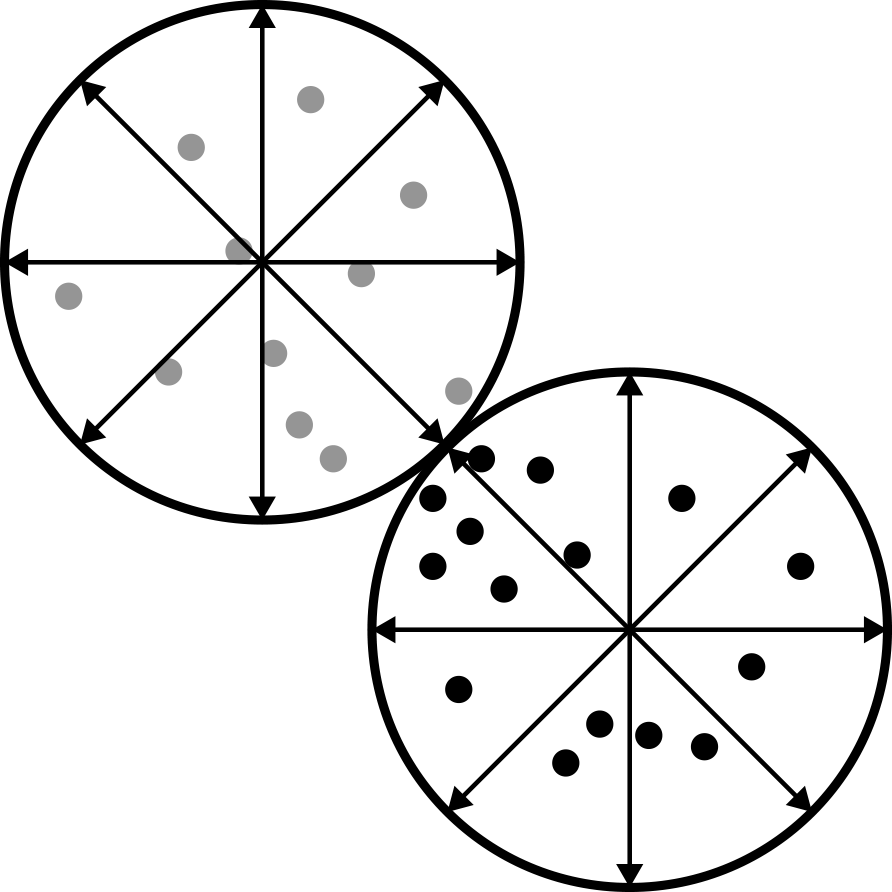
Figure 9-2. In a lot of ways, K-Means resembles a pizza
To construct K-Means clusters we need to have some sort of measurement for distance from the center. Back in Chapter 3 we introduced a few distance metrics, such as:
- Manhattan distance
-
- Euclidean distance
-
- Minkowski distance
-
- Mahalanobis distance
-
For a refresher on the metrics discussed here, review K-Nearest Neighbors in Chapter 3.
Downside of K-Means Clustering
One drawback of this procedure is that everything must have a hard boundary. This means that a data point can only be in one cluster and not straddle the line between two of them. K-Means also prefers spherical data since most of the time the Euclidean distance is being used. When looking at a graphic like Figure 9-3, where the data in the middle could go either direction (to a cluster on the left or right), the downsides become obvious.
EM Clustering
Instead of focusing on finding a centroid and then data points that relate to it, EM clustering focuses on solving a different problem. Let’s say that you want to split your data points into either cluster 1 or 2. You want a good guess of whether the data is in either cluster but don’t care if there’s some fuzziness. Instead of an exact assignment, we really want a probability that the data point is in each cluster.
Another way to think of clustering is how we interpret things like music. Classically speaking, Bach is Baroque music, Mozart is classical, and Brahms is Romantic. Using an algorithm like K-Means would probably work well for classical music, but for more modern music things aren’t that simple. For instance, jazz is extremely nuanced. Miles Davis, Sun Ra, and others really don’t fit into a categorization. They were a mix of a lot of influences.
So instead of classifying music like jazz we could take a more holistic approach through EM clustering. Imagine we had a simple example where we wanted to classify our jazz collection into either fusion or swing. It’s a simplistic model, but we could start out with the assumption that music could be either swing or fusion with a 50% chance. Notating this using math, we could say that zk = < 0.5, 0.5 >. Then if we were to run a special algorithm to determine what “Bitches Brew—Miles Davis” was in, we might find zk = < 0.9, 0.1 > or that it’s 90% fusion. Similarly if we were to run this on something like “Oleo—Sonny Rollins” we might find the opposite to be true with 95% being swing.
The beauty of this kind of thinking is that in practice, data doesn’t always fit into a category. But how would an algorithm like this work if we were to write it?

Figure 9-3. This shows how clusters can actually be much softer
Algorithm
The EM clustering algorithm is an iterative process to converge on a cluster mapping. It completes two steps in each iteration: expect and maximize.
But what does that mean? Expectation and maximization could mean a lot.
Expectation
Expectation is about updating the truth of the model and seeing how well we are mapping. In a lot of ways this is a test-driven approach to building clusters—we’re figuring out how well our model is tracking the data. Mathematically speaking, we estimate the probability vector for each row of data given its previous value.
On first iteration we just assume that everything is equal (unless you have some domain knowledge you feed into the model). Given that information we calculate the log likelihood of theta in the conditional distribution between our model and the true value of the data. Notated it is:
θ is the probability model we have assigned to rows. Z and X are the distributions for our cluster mappings and the original data points.
Maximization
Just estimating the log likelihood of something doesn’t solve our problem of assigning new probabilities to the Z distribution. For that we simply take the argument max of the expectation function. Namely, we are looking for the new θ that will maximize the log likelihood:
The unfortunate thing about EM clustering is that it does not necessarily converge and can falter when mapping data with singular covariances. We will delve into more of the issues related with EM clustering in “Example: Categorizing Music”. First we need to talk about one thing that all clustering algorithms share in common: the impossibility theorem.
The Impossibility Theorem
There is no such thing as free lunch and clustering is no exception. The benefit we get out of clustering to magically map data points to particular groupings comes at a cost. This was described by Jon Kleinberg, who touts it as the impossibility theorem, which states that you can never have more than two of the following when clustering:
-
Richness
-
Scale invariance
-
Consistency
Richness is the notion that there exists a distance function that will yield all different types of partitions. What this means intuitively is that a clustering algorithm has the ability to create all types of mappings from data points to cluster assignments.
Scale invariance is simple to understand. Imagine that you were building a rocket ship and started calculating everything in kilometers until your boss said that you need to use miles instead. There wouldn’t be a problem switching; you just need to divide by a constant on all your measurements. It is scale invariant. Basically if the numbers are all multiplied by 20, then the same cluster should happen.
Consistency is more subtle. Similar to scale invariance, if we shrank the distance between points inside of a cluster and then expanded them, the cluster should yield the same result. At this point you probably understand that clustering isn’t as good as many originally think. It has a lot of issues and consistency is definitely one of those that should be called out.
For our purposes K-Means and EM clustering satisfy richness and scale invariance but not consistency. This fact makes testing clustering just about impossible. The only way we really can test is by anecdote and example, but that is okay for analysis purposes.
In the next section we will analyze jazz music using K-Means and EM clustering.
Example: Categorizing Music
Music has a deep history of recordings and composed pieces. It would take an entire degree and extensive study of musicology just to be able to effectively categorize it all.
The ways we can sort music into categories is endless. Personally I sort my own record collection by artist name, but sometimes artists will perform with one another. On top of that, sometimes we can categorize based on genre. Yet what about the fact that genres are broad—such as jazz, for instance? According to the Montreux Jazz Festival, jazz is anything you can improvise over. How can we effectively build a library of music where we can divide our collection into similar pieces of work?
Instead let’s approach this by using K-Means and EM clustering. This would give us a soft clustering of music pieces that we could use to build a taxonomy of music.
In this section we will first determine where we will get our data from and what sort of attributes we can extract, then determine what we can validate upon. We will also discuss why clustering sounds great in theory but in practice doesn’t give us much except for clusters.
Setup Notes
All of the code we’re using for this example can be found on GitHub.
Python is constantly changing so the README is the best place to come up to speed on running the examples.
Gathering the Data
There is a massive amount of data on music from the 1100s through today. We have MP3s, CDs, vinyl records, and written music. Without trying to classify the entire world of music, let’s determine a small subsection that we can use. Since I don’t want to engage in any copyright suits we will only use public information on albums. This would be Artist, Song Name, Genre (if available), and any other characteristics we can find. To achieve this we have access to a plethora of information contained in Discogs.com. They offer many XML data dumps of records and songs.
Also, since we’re not trying to cluster the entire data set of albums in the world, let’s just focus on jazz. Most people would agree that jazz is a genre that is hard to really classify into any category. It could be fusion, or it could be steel drums.
To get our data set I downloaded metadata (year, artist, genre, etc.) for the best jazz albums (according to http://www.scaruffi.com/jazz/best100.html). The data goes back to 1940 and well into the 2000s. In total I was able to download metadata for about 1,200 unique records. All great albums!
But that isn’t enough information. On top of that I annotated the information by using the Discogs API to determine the style of jazz music in each.
After annotating the original data set I found that there are 128 unique styles associated with jazz (at least according to Discogs). They range from aboriginal to vocal.
Coding Design
Although this chapter uses two different algorithms (EM clustering and K-Means clustering), the code will focus on EM clustering and will follow the data flow in Figure 9-4.
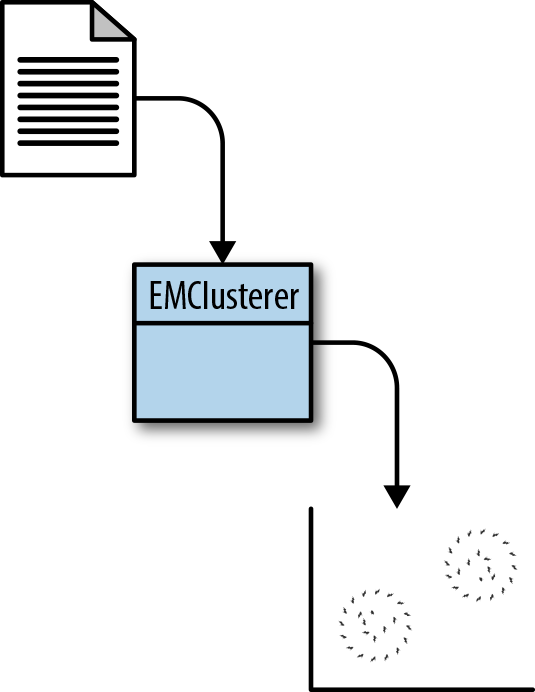
Figure 9-4. EM clustering class
Analyzing the Data with K-Means
Like we did with KNN, we need to figure out an optimal K. Unfortunately with clustering there really isn’t much we can test with except to just see whether we split into two different clusters.
But let’s say that we want to fit all of our records on a shelf and have 25 slots—similar to the IKEA bookshelf. We could run a clustering of all of our data using K = 25.
importcsvfromsklearn.clusterimportKMeansdata=[]artists=[]years=[]withopen('data/annotated_jazz_albums.csv','r')ascsvfile:reader=csv.DictReader(csvfile)headers=reader.fieldnames[3:]forrowinreader:artists.append(row['artist_album'])years.append(row['year'])data.append([int(row[key])forkeyinheaders])clusters=KMeans(n_clusters=25).fit_predict(data)withopen('data/clustered_kmeans.csv','w')ascsvfile:fieldnames=['artist_album','year','cluster']writer=csv.DictWriter(csvfile,fieldnames=fieldnames)writer.writeheader()fori,clusterinenumerate(clusters):writer.writerow({'artist_album':artists[i],'year':years[i],'cluster':cluster})
That’s it! Of course clustering without looking at what this actually tells us is useless. This does split the data into 25 different categories, but what does it all mean?
Looking at a graphic of year versus assigned cluster number yields interesting results (Figure 9-5).
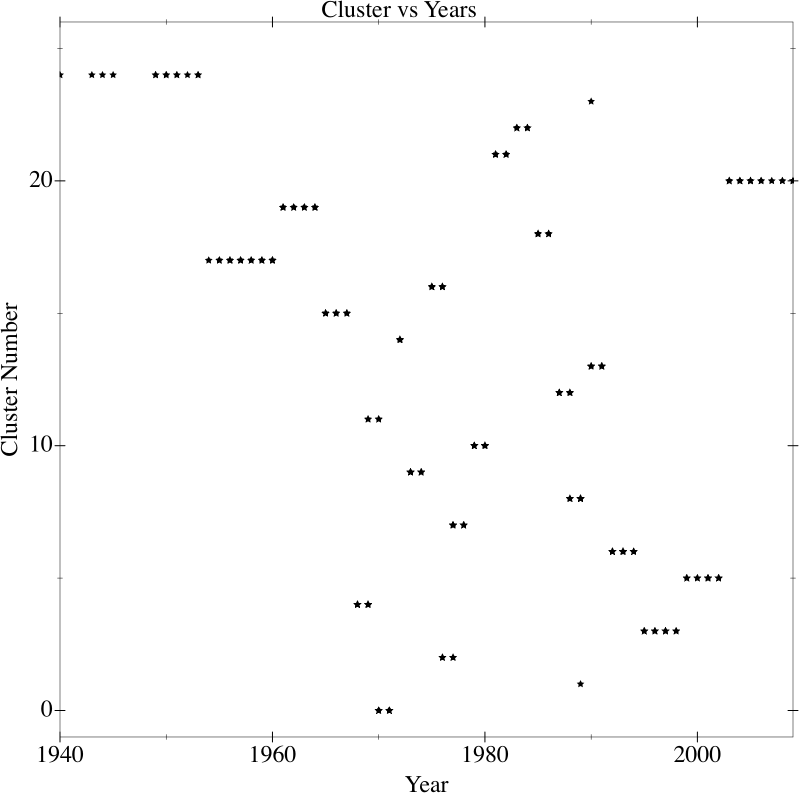
Figure 9-5. K-Means applied to jazz albums
As you can see, jazz starts out in the Big Band era pretty much in the same cluster, transitions into cool jazz, and then around 1959 it starts to go everywhere until about 1990 when things cool down a bit. What’s fascinating is how well that syncs up with jazz history.
EM Clustering Our Data
With EM clustering, remember that we are probabilistically assigning to different clusters—it isn’t 100% one or the other. This could be highly useful for our purposes since jazz has so much crossover.
Let’s go through the process of writing our own code and then use it to map the same data that we have from our jazz data set, then see what happens.
Our first step is to be able to initialize the cluster. If you remember we need to have indicator variables zt that follow a uniform distribution. These tell us the probability that each data point is in each cluster.
Stepping back into our example, we first need to write a helper function that returns the density of a multivariate normal distribution. This is based on how R does this:
fromcollectionsimportnamedtupleimportrandomimportloggingimportmathimportnumpyasnpfromnumpy.linalgimportLinAlgErrordefdvmnorm(x,mean,covariance,log=False):"""density function for the multivariate normal distributionbased on sources of R library 'mvtnorm':rtype : np.array:param x: vector or matrix of quantiles. If x is a matrix, each row is takento be a quantile:param mean: mean vector, np.array:param covariance: covariance matrix, np.array:param log: if True, densities d are given as log(d), default is False"""n=covariance.shape[0]try:dec=np.linalg.cholesky(covariance)exceptLinAlgError:dec=np.linalg.cholesky(covariance+np.eye(covariance.shape[0])*0.0001)tmp=np.linalg.solve(dec,np.transpose(x-mean))rss=np.sum(tmp*tmp,axis=0)logretval=-np.sum(np.log(np.diag(dec)))-0.5*n*np.log(2*math.pi)-0.5*rssiflog:returnlogretvalelse:returnnp.exp(logretval)
Using all this setup we can now build an EMClustering class that will do our EM clustering and log outputs as needed.
This class has the following methods of note:
partitions-
Will return the cluster mappings of the data if they are set.
data-
Will return the data object passed in.
labels-
Will return the membership weights for each cluster.
clusters-
Will return the clusters.
setup-
This does all of the setup for the EM clustering.
classEMClustering(object):logger=logging.getLogger(__name__)ch=logging.StreamHandler()formatstring='%(asctime)s-%(name)s-%(levelname)s-%(message)s'formatter=logging.Formatter(formatstring)ch.setFormatter(formatter)logger.addHandler(ch)logger.setLevel(logging.DEBUG)cluster=namedtuple('cluster','weight, mean, covariance')def__init__(self,n_clusters):self._data=Noneself._clusters=Noneself._membership_weights=Noneself._partitions=Noneself._n_clusters=n_clusters@propertydefpartitions(self):returnself._partitions@propertydefdata(self):returnself._data@propertydeflabels(self):returnself._membership_weights@propertydefclusters(self):returnself._clustersdefsetup(self,data):self._n_samples,self._n_features=data.shapeself._data=dataself._membership_weights=np.ones((self._n_samples,self._n_clusters))/self._n_clustersself._s=0.2indices=list(range(data.shape[0]))random.shuffle(indices)pick_k_random_indices=random.sample(indices,self._n_clusters)self._clusters=[]forcluster_numinrange(self._n_clusters):mean=data[pick_k_random_indices[cluster_num],:]covariance=self._s*np.eye(self._n_features)mapping=self.cluster(1.0/self._n_clusters,mean,covariance)self._clusters.append(mapping)self._partitions=np.empty(self._n_samples,dtype=np.int32)
At this point we have set up all of our base case stuff. We have @k, which is the number of clusters, @data is the data we pass in that we want to cluster, @labels are an array full of the probabilities that the row is in each cluster, and @classes holds on to an array of means and covariances, which tells us where the distribution of data is. Last, @partitions are the assignments of each data row to cluster index.
Now we need to build our expectation step, which is to figure out what the probability of each data row is in each cluster. To do this we need to write a new method, expect, which will do this:
classEMClustering(object):# __init__# setup()defexpect(self):log_likelyhood=0forcluster_num,clusterinenumerate(self._clusters):log_density=dvmnorm(self._data,cluster.mean,cluster.covariance,log=True)membership_weights=cluster.weight*np.exp(log_density)log_likelyhood+=sum(log_density*self._membership_weights[:,cluster_num])self._membership_weights[:,cluster_num]=membership_weightsforsample_num,probabilitiesinenumerate(self._membership_weights):prob_sum=sum(probabilities)self._partitions[sample_num]=np.argmax(probabilities)ifprob_sum==0:self._membership_weights[sample_num,:]=np.ones_like(probabilities)/self._n_clusterselse:self._membership_weights[sample_num,:]=probabilities/prob_sumself.logger.debug('log likelyhood%f',log_likelyhood)
The first part of this code iterates through all classes, which holds on to the means and covariances of each cluster. From there we want to find the inverse covariance matrix and the determinant of the covariance. For each row we calculate a value that is proportional to the probability that the row is in a cluster. This is:
This is effectively a Gaussian distance metric to help us determine how far outside of the mean our data is.
Let’s say that the row vector is exactly the mean. This equation would reduce down to pij = det(C), which is just the determinant of the covariance matrix. This is actually the highest value you can get out of this function. If for instance the row vector was far away from the mean vector, then pij would become smaller and smaller due to the exponentiation and negative fraction in the front.
The nice thing is that this is proportional to the Gaussian probability that the row vector is in the mean. Since this is proportional and not equal to, in the last part we end up normalizing to sum up to 1.
Now we can move on to the maximization step:
classEMClustering(object):# __init__# setup()# expectdefmaximize(self):forcluster_num,clusterinenumerate(self._clusters):weights=self._membership_weights[:,cluster_num]weight=np.average(weights)mean=np.average(self._data,axis=0,weights=weights)covariance=np.cov(self._data,rowvar=False,ddof=0,aweights=weights)self._clusters[cluster_num]=self.cluster(weight,mean,covariance)
Again here we are iterating over the clusters called @classes. We first make an array called sum that holds on to the weighted sum of the data happening. From there we normalize to build a weighted mean. To calculate the covariance matrix we start with a zero matrix that is square and the width of our clusters. Then we iterate through all row vectors and incrementally add on the weighted difference of the row and the mean for each combination of the matrix. Again at this point we normalize and store.
Now we can get down to using this. To do that we add two convenience methods that help in querying the data:
classEMClustering(object):# __init__# setup# expect# maximizedeffit_predict(self,data,iteration=5):self.setup(data)foriinrange(iteration):self.logger.debug('Iteration%d',i)self.expect()self.maximize()returnself
The Results from the EM Jazz Clustering
Back to our results of EM clustering with our jazz music. To actually run the analysis we run the following script:
importcsvimportnumpyasnpfromem_clusteringimportEMClusteringnp.set_printoptions(threshold=9000)data=[]artists=[]years=[]# with open('data/less_covariance_jazz_albums.csv', 'rb') as csvfile:withopen('data/annotated_jazz_albums.csv','r')ascsvfile:reader=csv.DictReader(csvfile)headers=reader.fieldnames[3:]forrowinreader:artists.append(row['artist_album'])years.append(row['year'])data.append([int(row[key])forkeyinheaders])clusterer=EMClustering(n_clusters=25)clusters=clusterer.fit_predict(np.array(data))(clusters.partitions)
The first thing you’ll notice about EM clustering is that it’s slow. It’s not as quick as calculating new centroids and iterating. It has to calculate covariances and means, which are all inefficient. Ockham’s Razor would tell us here that EM clustering is most likely not a good use for clustering big amounts of data.
The other thing that you’ll notice is that our annotated jazz music will not work because the covariance matrix of this is singular. This is not a good thing; as a matter of fact this problem is ill suited for EM clustering because of this, so we have to transform it into a different problem altogether.
We do that by collapsing the dimensions into the top two genres by index:
importcsvwithopen('data/less_covariance_jazz_albums.csv','w')ascsvout:writer=csv.writer(csvout,delimiter=',')# Write the header of the CSV filewriter.writerow(['artist_album','key_index','year','Genre_1','Genre_2'])withopen('data/annotated_jazz_albums.csv','r')ascsvin:reader=csv.DictReader(csvin)forrowinreader:genre_count=0genres=[0,0]genre_idx=0idx=0forkey,valueinrow.items():breakifgenre_idx==2ifvalue=='1':genres[genre_idx]=idxgenre_idx+=1idx+=1ifgenres[0]>0||genres[1]>0:line=[row['artist_album'],row['key_index'],row['year'],genres[0],genres[1]]writer.writerow(line)
Basically what we are doing here is saying for the first two genres, let’s assign a genre index to it and store it. We’ll skip any albums with zero information assigned to them.
At this point we are able to run the EM clustering algorithm, except that things become too difficult to actually cluster. This is an important lesson with EM clustering. The data we have actually doesn’t cluster because the matrix has become too unstable to invert.
Some possibilities for refinement would be to try out K-Means or other clustering algorithms, but really a better approach would be to work on the data some more. Jazz albums are a fun example but data-wise aren’t very illustrative. We could, for instance, expand using some more musical genomes, or feed this into some sort of text-based model. Or maybe we could spelunk for musical queues using fast Fourier transforms! The possibilities are really endless but this gives us a good start.
Conclusion
Clustering is useful but can be a bit hard to control since it is unsupervised. Add the fact that we are dealing with the impossibility of having consistency, richness, and scale-invariance all at once and clustering can be a bit useless in many contexts. But don’t let that get you down—clustering can be useful for analyzing data sets and splitting data into arbitrary clusters. If you don’t care how they are split and just want them split up, then clustering is good. Just know that there are sometimes odd circumstances.
