134 Zero to Genetic Engineering Hero - Chapter 5 - Extracting your engineered proteins
•
Ribosome: The ribosome is a hybrid of protein
and RNA, and it catalyzes the chemical reactions
for joining amino acids together. The ribosome is
analogous to the RNA polymerase in transcription.
•
rRNA: Ribosomal RNA (rRNA) is embedded in the
ribosome protein and helps it to lock onto the RNA
transcript’s ribosomal binding site (RBS) just prior
to when translation starts. It does this by comple-
menting (zippers with) the ribonucleotides that
make up the RBS.
•
tRNA: Transfer RNA (tRNA) are specialized “go-be-
tweens” that complement the RNA transcript
coding region and have an amino acid bound to its
other end. They bridge the gap between RNA and
amino acids.
A tRNA with an amino acid attached to it is called
an aminoacyl-tRNA (Figure 5-15), but they are
commonly referred to just as tRNAs.
During translation, the ribosome “reads” three ribo-
nucleotides from the RNA transcript at one time
using tRNA. It does this because each tRNA antico-
don complements three ribonucleotides in the RNA
transcript coding region at once. A group of three
ribonucleotides in an RNA transcript are called a
codon (Figure 5-15). In other words, the three letter
codon of the RNA sequence (e.g., AUG) complements
the anticodon sequence of a tRNA (e.g., UAC). Similar
to how during transcription nucleotides bump around
the cell until the right one bumps its way into the RNA
polymerase and becomes attached to the growing
transcript, tRNAs bump around the cell and when
the right tRNA bumps into a ribosome that is loaded
with mRNA and the anticodon complements all three
ribonucleotides in the codon, the ribosome adds the
amino acid on that tRNA to a growing chain of amino
acids (Figure 5-16 and Figure 5-17). In other words, it
takes three RNA nucleotides of information to encode
one amino acid.
Each amino acid is encoded by one or more RNA
codons (Table 5-2). You’ll see in Table 5-2 that there
are 64 possible RNA codons. Proteins are typically
chains of 50 amino acids or more which require 50
codons or more. This means that the RNA transcripts
needs to be at least 150 ribonucleotides, plus the
non-coding RBS and terminator. This further means
that the DNA gene must be 150 nucleotides, plus
promoter (for transcription), the RBS (for translation),
and the terminator (for transcription)! The RNA codon
table (Table 5-2) is used to know which RNA codons
code for each amino acid. Note that different codons
can code for the same amino acid. For example, C-A-U
and C-A-C both encode for the amino acid histidine.
This means that there is an aminoacyl-tRNA (tRNA)
that, at one end, has an RNA anticodon sequence that
complements the C-A-C or C-A-U, and has a histidine
amino acid at the other end.
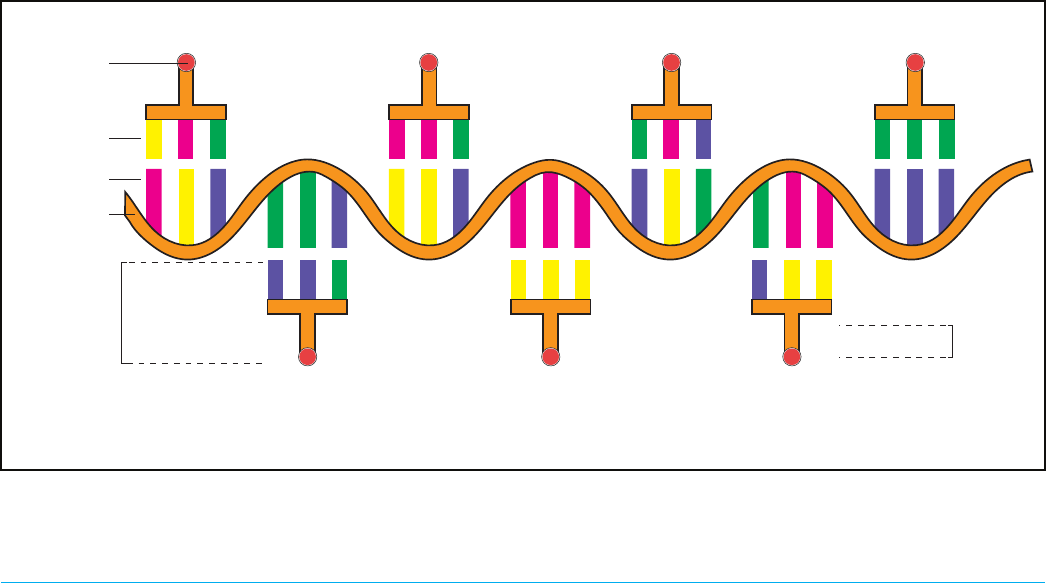
Figure 5-15. tRNAs are the physical connectors that bind to the RNA codons in the ribosome. When the appropriate tRNA binds to a
codon (and they complement), the amino acid at the “base of the T” is added to the growing amino acid chain.
RNA transcript
Codon
Anti-Codon
Amino acid
aminoacyl-tRNA
(tRNA)
Amino acids
(Proteins)
U A C
G G C
Amino acid 2
C C G
A A C
U U U
Amino acid 4 Amino acid 6
A A A
C A G
G U U
C C C
C A A
Amino acid 1
A U G
Amino acid 3
U U G
Amino acid 5
G U C G G G
Amino acid 7
Book _genetic engineering hero-AUG2021.indb 134Book _genetic engineering hero-AUG2021.indb 134 8/18/21 12:03 PM8/18/21 12:03 PM

135Zero to Genetic Engineering Hero - Chapter 5 - Extracting your engineered proteins
tRNA Going Deeper 5-4
In Figure 5-15, you can see the tRNAs as “T” shaped, with an amino acid at the bottom and three ribonucle-
otides at the top. While it is true that in their real physical form the tRNAs are somewhat T shaped (Figure
5-14), the tRNAs of Figure 5-15 are a cartoon representation of the 3:1 ratio, and not faithful to that physical
shape. Visit the Protein Data Bank webpage to see more in-depth information about tRNAs:
https://pdb101.rcsb.org/motm/15
Figure 5-16. As the ribosome travels downstream, it “reads” the RNA transcript. A bumping tRNA that is complementary to the RNA
codon in the ribosome will bind strongly, and this triggers the ribosome to attach the amino acid end to the growing amino acid pep-
tide strand. If the three ribonucleotides (codon) and tRNA anticodon do not bind strongly, the tRNA will bump out of the ribosome,
and eventually the correct tRNA will bump into the ribosome and be added to the growing string of amino acids.
C A A
Ribosome
rRNA
tRNA
U U G
C A A
Amino Acid Chain
RNA transcript
Figure 5-17. Comparing the complementarity of DNA, RNA, and tRNAs.
RNA transcriptDNA
tRNA
DNADNA RNA transcript
T A C G C A
A U G C G U
A U G C G U
TACGCA
ATGCGT
U A C G C A
Book _genetic engineering hero-AUG2021.indb 135Book _genetic engineering hero-AUG2021.indb 135 8/18/21 12:03 PM8/18/21 12:03 PM

136 Zero to Genetic Engineering Hero - Chapter 5 - Extracting your engineered proteins
Table 5-2. Translation cipher - RNA codon table
1st base 2nd base 3rd base
U C A G
U
UUU
F/Phe
Phenylalanine
UCU
S/Ser
Serine
UAU
Y/Tyr
Tyrosine
UGU
C/Cys
Cysteine
U
UUC UCC UAC UGC C
UUA
L/Leu
Leucine
UCA UAA Stop (Ochre) UGA Stop (Opal) A
UUG UCG UAG Stop (Amber) UGG
W/Trp Trypto-
phan
G
C
CUU CCU
P/Pro
Proline
CAU
H/His
Histidine
CGU
R/Arg
Arginine
U
CUC CCC CAC CGC C
CUA CCA CAA
G/Gln
Glutamine
CGA A
CUG CCG CAG CGG G
A
AUU
I/Ile
Isoleucine
ACU
T/Thr
Theronine
AAU
N/Asn
Asparagine
AGU
S/Ser
Serine
U
AUC ACC AAC AGC C
AUA ACA AAA
K/Lys
Lysine
AGA
R/Arg
Arginine
A
AUG
M/Met/Methi-
onine
ACG AAG AGG G
G
GUU
V/Val
Valine
GCU
A/Ala
Alanine
GAU
D/Asp
Aspartic acid
(Aspartate)
GGU
G/Gly
Glycine
U
GUC GCC GAC GGC C
GUA GCA GAA
E/Glu
Glutamic acid
(Glutamate)
GGA A
GUG GCG GAG GGG G
Four B’s for Choosing Correct Amino Acid: Going Deeper 5-5
It should be emphasized that similar to RNA polymerase in transcription, the ribosome does not have the
“intelligence” to read, analyse, and understand the RNA sequence and then use that information to decide
which amino acid to attach to the growing chain. Rather the Four B’s are at the centre of it all! Recall the Four
B’s concept you read about at the beginning of the Fundamentals in Chapter 4: Bump, Bind, Burst (optional),
and Bump.
In the case of the ribosome, there are many tRNAs with amino acids attached to them that are produced
by the cell and are bumping around inside the cell. This pool or tRNAs is also capable of bumping into and
out of the ribosome. As a ribosome is positioned at a particular spot on the RNA, three ribonucleotides are
available to bind to the correct tRNA. tRNA bump in and out of the ribosome, and when the correct tRNA
that has an anti-codon that matches and binds to the RNA codon, this triggers the ribosome to complete
the next step of attaching the amino acid to the growing amino acid chain (protein).
So while the ribosome doesn’t have human-like intelligence and decision making skills, the “information”
that determines which aminoacyl-tRNA is correctly chosen is embedded within the system in the form
hydrogen bonding between the ribonucleotides of the codon and anti-codon! In other words, the informa-
tion is “hardwired into the system”.
Book _genetic engineering hero-AUG2021.indb 136Book _genetic engineering hero-AUG2021.indb 136 8/18/21 12:03 PM8/18/21 12:03 PM

137Zero to Genetic Engineering Hero - Chapter 5 - Extracting your engineered proteins
During Translation: Locating the
starting point for translation
How does the ribosome know precisely where to start
translating the coding region of the RNA transcript?
You’ve already learned that the initiation factors 1,
2, and 3 are responsible for recruiting the ribosome
and the RBS at the 5’ end of the RNA transcript, and
the rRNA in the ribosome lock it into place. Let’s now
learn how, once the ribosome is positioned and ready
to go, it knows exactly where to start.
While it may seem like the RBS would be the starting
point for translation, the RBS is actually an approx-
imation of the start point and only serves to lock in
the ribosome close to the start point. This is similar
to typing - when you start typing, your hands will
hover over the keyboard, with your fingers near,
but not on the keys you will soon be using - you are
locked into your keyboard but not yet using it. In the
RNA transcript, the starting point is specic: a single
codon from the 64 possible RNA codons is called
Start Codon, and it is required to be near the RBS for
the ribosome to start translating. The start codon is
also the same codon that encodes for the amino acid
methionine. Have a look in Table 5-2, what is the
three-letter RNA codon for Methionine/Met?
A-U-G! The start codon will be found 2 to 6 ribonu-
cleotides downstream of the ribosomal binding site
(Figure 5-18). Once the ribosome binds to the ribo-
somal binding site, the initiation factors, which are
also bound to a special “starter” methionine tRNA
called fMet (formylmethionine), help kick-start the
translation process. The initiation factors “give” the
fMet to the ribosome to become the rst amino acid
of the protein and then the initiation factors leave.
Only for the A-U-G start codon is the fMet used and
any A-U-G codon that is present inside of a sequence
being translated, uses the regular methionine tRNA.
This is in part because once translation starts, the
initiation factors leave the ribosome and because the
initiation factors have the unique ability to bind the
fMet and give it to the ribosome during translation
initiation, fMet doesn’t usually play a role during the
rest of translation.
Protein Data Bank (PDB) of Ribosome Web Search Breakout
The Protein Data Bank (PDB) is an online database of X-ray crystal structures of biomolecules. There is a
fantastic overview of ribosomes and how they work. https://pdb101.rcsb.org/motm/121
Be the cell Machinery! DNA to Protein Breakout Exercise
In this super quick breakout exercise, you’re going to practice what you know about how a cell is able to “read” DNA
and ultimately create protein.
Table 5-3. Practice the transcription and translation ciphers
DNA + leading strand AGG GAG GCC GAT
DNA- template strand
RNA (codon)
tRNA (anti-codon)
Amino acid (protein)
*Remember that in RNA there are no “T’s”! Refer back to Table 4-2
Book _genetic engineering hero-AUG2021.indb 137Book _genetic engineering hero-AUG2021.indb 137 8/18/21 12:03 PM8/18/21 12:03 PM
138 Zero to Genetic Engineering Hero - Chapter 5 - Extracting your engineered proteins
After the start codon, the ribosome continues to move
along the coding region of the RNA transcript (Figure
5-18). Thousands of the 64 different tRNA molecules
complete the Four B’s of basic cell operation and,
when the correct tRNA enters a translating ribo-
some and its anticodon matches perfectly with the
RNA transcript codon present in the ribosome, the
ribosome takes and connects the amino acid to the
growing chain (Figure 5-16). As new tRNA come into
the ribosome, it releases the previous tRNA that lost
its amino acid to the growing protein chain.
As the ribosome is translating the amino acid chain,
the amino acids in the chain begin to interact with
one another. The 20 amino acids are each quite
unique - some are negatively charged, some posi-
tively charged, and some not charged (Figure 3-30).
Some of these have sizeable bulky side chains (argi-
nine), and some have small side chains (glycine). It
is this wide variety of amino acid side chains that
gives the amino acids different “personalities”. As
the amino acid chain gets longer during translation,
the different amino acids in the chain bump and bind
or repel each other to form a complex and beautiful
three-dimensional shape (Figure 3-28). This three-
dimensional shape of proteins is what results in their
function - like being colorful!
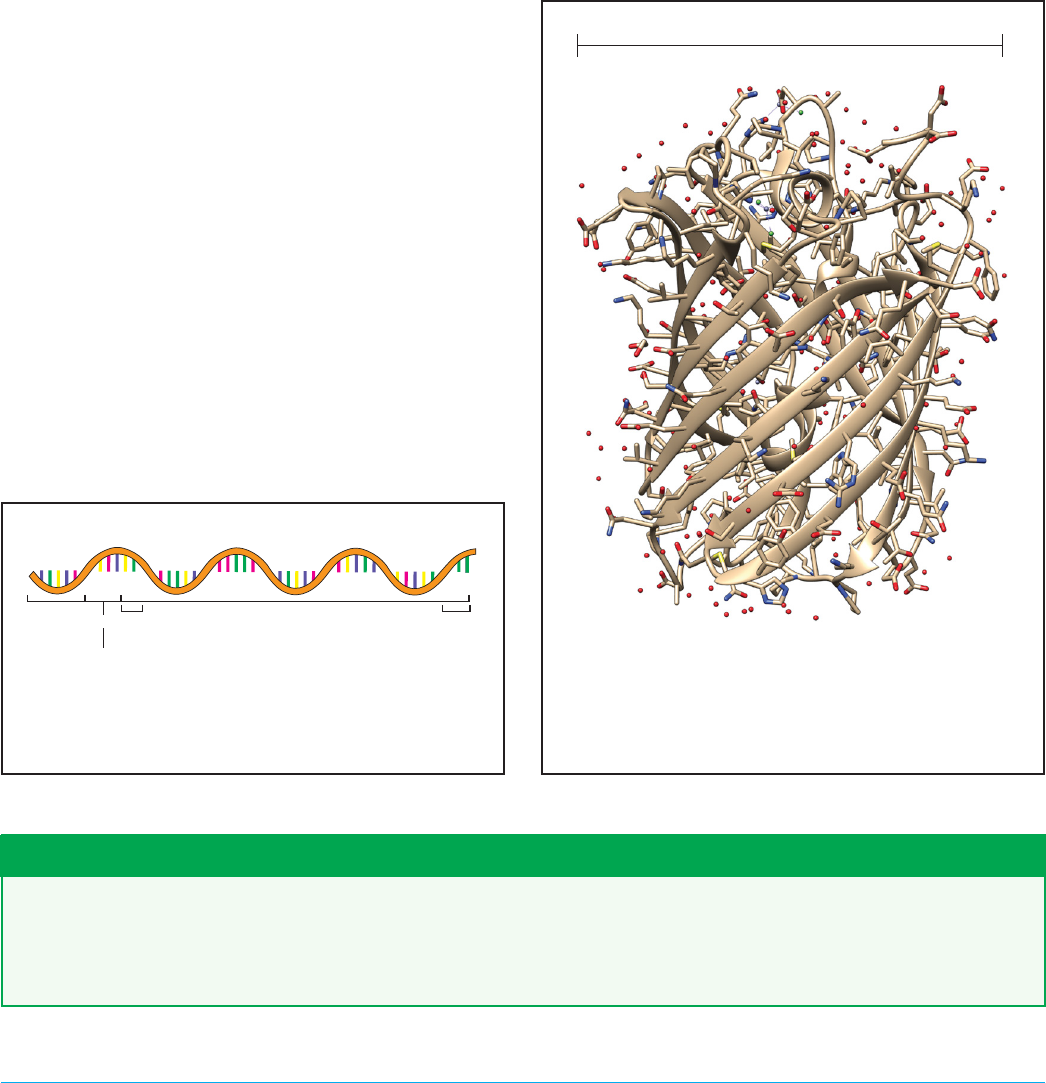
In Figure 5-19, you will find one product of trans-
lation, the crystal structure of a protein called Red
Fluorescent Protein (RFP). Just like the structure of
DNA you saw in Chapter 1 (Figure 1-19), you will see
the red, blue, and gray segments of this structure
which correspond to oxygen, nitrogen, and carbon,
respectively.
In Figure 5-19 you will also see a “ribbon” throughout
the structure, and this is commonly used as a guide to
help visualize the intricate and beautiful “beta barrel”
structure of the protein but is not a physical part of
the protein. RFP is only one example of thousands
of proteins that E. coli make, all of which have differ-
ent 3D shapes. You’ll see that RFP is very small at 3.5
nanometers wide. Compared to a DNA helix at ~12
nanometers wide, proteins can be quite small.
Figure 5-19. A protein is a long chain of amino acids folded
up into a three-dimensional structure. In this example, red
uorescent protein (RFP) has a beta-barrel structure. RFP
is a small protein that is about 250 amino acids long and is
about 3.5 nanometers wide. Crystal structure data source:
RCSB PDB: 2VAD.
3.5 nanometers (0.0000000035 m)
Figure 5-18. RNA transcripts are transcribed by RNA poly-
merase and are made up of ribonucleotides. A transcript has
a ribosomal binding site (RBS), 2-6 nucleotide spacer, start
codon, protein coding region, and a stop codon.
RNA Transcript
RBS
2-6 nt space
Stop
Codon
Protein coding region
5’
3’
Start Codon
The methionine start codon Going Deeper 5-6
So does this mean that every protein starts with methionine? Yes, it does, at least when it is made initially.
E. coli cells have some protein enzymes called proteases that are able to clip off the ends of proteins after
translation. Typically, proteases will cut off several amino acids from either end of the protein, one of which
could be the starter methionine.
Book _genetic engineering hero-AUG2021.indb 138Book _genetic engineering hero-AUG2021.indb 138 8/18/21 12:03 PM8/18/21 12:03 PM
..................Content has been hidden....................
You can't read the all page of ebook, please click here login for view all page.
