2 The mathematical building blocks of neural networks
This chapter covers
- A first example of a neural network
- Tensors and tensor operations
- How neural networks learn via backpropagation and gradient descent
Understanding deep learning requires familiarity with many simple mathematical concepts: tensors, tensor operations, differentiation, gradient descent, and so on. Our goal in this chapter will be to build up your intuition about these notions without getting overly technical. In particular, we’ll steer away from mathematical notation, which can introduce unnecessary barriers for those without any mathematics background and isn’t necessary to explain things well. The most precise, unambiguous description of a mathematical operation is its executable code.
To provide sufficient context for introducing tensors and gradient descent, we’ll begin the chapter with a practical example of a neural network. Then we’ll go over every new concept that’s been introduced, point by point. Keep in mind that these concepts will be essential for you to understand the practical examples in the following chapters.
After reading this chapter, you’ll have an intuitive understanding of the mathematical theory behind deep learning, and you’ll be ready to start diving into Keras and TensorFlow in chapter 3.
2.1 A first look at a neural network
Let’s look at a concrete example of a neural network that uses Keras to learn to classify handwritten digits. Unless you already have experience with Keras or similar libraries, you won’t understand everything about this first example right away. That’s fine. In the next chapter, we’ll review each element in the example and explain them in detail. So don’t worry if some steps seem arbitrary or look like magic to you—we’ve got to start somewhere.
The problem we’re trying to solve here is to classify grayscale images of handwritten digits (28 × 28 pixels) into their 10 categories (0 through 9). We’ll use the MNIST dataset, a classic in the machine learning community, which has been around almost as long as the field itself and has been intensively studied. It’s a set of 60,000 training images, plus 10,000 test images, assembled by the National Institute of Standards and Technology (the NIST in MNIST) in the 1980s. You can think of “solving” MNIST as the “Hello World” of deep learning—it’s what you do to verify that your algorithms are working as expected. As you become a machine learning practitioner, you’ll see MNIST come up over and over again in scientific papers, blog posts, and so on. You can see some MNIST samples in figure 2.1.

Figure 2.1 MNIST sample digits
In machine learning, a category in a classification problem is called a class. Data points are called samples. The class associated with a specific sample is called a label.
You don’t need to try to reproduce the example shown in the next code listing on your machine just now. If you wish to, you’ll first need to set up a deep learning workspace, which is covered in chapter 3. The MNIST dataset comes preloaded in Keras, in the form of a set of four R arrays, organized into two lists named train and test.
Listing 2.1 Loading the MNIST dataset in Keras
library(tensorflow)
library(keras)
mnist <- dataset_mnist()
train_images <- mnist$train$x
train_labels <- mnist$train$y
test_images <- mnist$test$x
test_labels <- mnist$test$y
train_images and train_labels form the training set, the data that the model will learn from. The model will then be tested on the test set, test_images and test_ labels. The images are encoded as R arrays, and the labels are an array of digits, ranging from 0 to 9. The images and labels have a one-to-one correspondence. Let’s look at the training data, shown here:
str(train_images)
int[1:60000, 1:28, 1:28] 0 0 0 0 0 0 0 0 0 0 …
str(train_labels)
int [1:60000(1d)] 5 0 4 1 9 2 1 3 1 4 …
And here’s the test data:
str(test_images)
int [1:10000, 1:28, 1:28] 0 0 0 0 0 0 0 0 0 0 …
str(test_labels)
int [1:10000(1d)] 7 2 1 0 4 1 4 9 5 9 …
The workflow will be as follows: first, we’ll feed the neural network the training data, train_images and train_labels. Then the network will learn to associate images and labels. Finally, we’ll ask the network to produce predictions for test_images, and we’ll verify whether these predictions match the labels from test_labels.
Let’s build the network, as shown in the next listing. Again, remember that you aren’t expected to understand everything about this example yet.
Listing 2.2 The network architecture
model <- keras_model_sequential(list(
layer_dense(units = 512, activation = "relu"),
layer_dense(units = 10, activation = "softmax")
))
The core building block of neural networks is the layer. You can think of a layer as a filter for data: some data goes in, and it comes out in a more useful form. Specifically, layers extract representations out of the data fed into them—hopefully, representations that are more meaningful for the problem at hand. Most of deep learning consists of chaining together simple layers that will implement a form of progressive data distillation. A deep learning model is like a sieve for data processing, made of a succession of increasingly refined data filters—the layers.
Here, our model consists of a sequence of two Dense layers, which are densely connected (also called fully connected) neural layers. The second (and last) layer is a 10-way softmax classification layer, which means it will return an array of 10 probability scores (summing to 1). Each score will be the probability that the current digit image belongs to one of our 10 digit classes.
To make the model ready for training, we need to pick the following three things as part of the compilation step, shown in listing 2.3:
- An optimizer—The mechanism through which the model will update itself based on the training data it sees, so as to improve its performance.
- A loss function—How the model will be able to measure its performance on the training data, and thus how it will be able to steer itself in the eright direction.
- Metrics to monitor during training and testing—Here, we care only about accuracy (the fraction of the images that were correctly classified).
The exact purpose of the loss function and the optimizer will be made clear throughout the next two chapters.
Listing 2.3 The compilation step
compile(model,
optimizer = "rmsprop",
loss = "sparse_categorical_crossentropy",
metrics = "accuracy")
Note that we don’t save the return value from compile() because the model is modified in place.
Before training, we’ll preprocess the data by reshaping it into the shape the model expects and scaling it so that all values are in the [0, 1] interval, as shown next. Previously, our training images were stored in an array of shape (60000, 28, 28) of type integer with values in the [0, 255] interval. We’ll transform it into a double array of shape (60000, 28 * 28) with values between 0 and 1.
Listing 2.4 Preparing the image data
train_images <- array_reshape(train_images, c(60000, 28 * 28))
train_images <- train_images / 255
test_images <- array_reshape(test_images, c(10000, 28 * 28))
test_images <- test_images / 255
Note that we use the array_reshape() function rather than the dim•() function to reshape the array. We’ll explain why later, when we talk about tensor reshaping.
We’re now ready to train the model, which in Keras is done via a call to the model’s fit() method—we fit the model to its training data.
Listing 2.5 “Fitting” the model
fit(model, train_images, train_labels, epochs = 5, batch_size = 128)
Epoch 1/5
60000/60000 [===========================] - 5s - loss: 0.2524 - acc:
![]() 0.9273
0.9273
Epoch 2/5
51328/60000 [=====================>.....] - ETA: 1s - loss: 0.1035 -
![]() acc: 0.9692
acc: 0.9692
Two quantities are displayed during training: the loss of the model over the training data, and the accuracy of the model over the training data. We quickly reach an accuracy of 0.989 (98.9%) on the training data.
Now that we have a trained model, we can use it to predict class probabilities for new digits—images that weren’t part of the training data, like those from the test set.
Listing 2.6 Using the model to make predictions
test_digits <- test_images[1:10, ]
predictions <- predict(model, test_digits)
str(predictions)
num [1:10, 1:10] 3.10e-09 3.53e-11 2.55e-07 1.00 8.54e-07 …
predictions[1, ]
[1] 3.103298e-09 1.175280e-10 1.060593e-06 4.761311e-05 4.189971e-12
[6] 4.062199e-08 5.244305e-16 9.999473e-01 2.753219e-07 3.826783e-06
Each number of index i in that array (predictions[1, ]) corresponds to the probability that digit image test_digits[1, ] belongs to class i. This first test digit has the highest probability score (0.9999473, almost 1) at index 8, so according to our model, it must be a 7 (because we start counting at 0):
which.max(predictions[1, ])
[1] 8
predictions[1, 8]
[1] 0.9999473
We can check that the test label agrees:
test_labels[1]
[1] 7
On average, how good is our model at classifying such never-before-seen digits? Let’s check by computing average accuracy over the entire test set.
Listing 2.7 Evaluating the model on new data
metrics <- evaluate(model, test_images, test_labels)
metrics["accuracy"]
accuracy
0.9795
The test set accuracy turns out to be 97.9%—that’s quite a bit lower than the training set accuracy (98.9%). This gap between training accuracy and test accuracy is an example of overfitting: the fact that machine learning models tend to perform worse on new data than on their training data. Overfitting is a central topic in chapter 3.
This concludes our first example. You just saw how you can build and train a neural network to classify handwritten digits in fewer than 15 lines of R code. In this chapter and the next, we’ll go into detail about every moving piece we just previewed and clarify what’s going on behind the scenes. You’ll learn about tensors, the data-storing objects going into the model; tensor operations, which layers are made of; and gradient descent, which allows your model to learn from its training examples.
2.2 Data representations for neural networks
In the previous example, we started from data stored in multidimensional arrays, also called tensors. In general, all current machine learning systems use tensors as their basic data structure. Tensors are fundamental to the field—so fundamental that TensorFlow was named after them. So, what’s a tensor?
At its core, a tensor is a container for data—usually numerical data—so, it’s a container for numbers. You may be already familiar with matrices, which are rank 2 tensors: tensors are a generalization of matrices to an arbitrary number of dimensions (note that in the context of tensors, a dimension is often called an axis).
R provides an implementation of tensors: array objects (constructed via base:: array()) are tensors. In this section we are focused on defining the concepts around tensors, so we will stick to using R arrays. Later in the book (chapter 3), we introduce another implementation of tensors (Tensorflow Tensors).
2.2.1 Scalars (rank 0 tensors)
A tensor that can contain only one number is called a scalar (or scalar tensor, or rank 0 tensor, or 0D tensor). R doesn’t have a data type to represent scalars (all numeric objects are vectors), but an R vector of length 1 is conceptually similar to a scalar.
2.2.2 Vectors (rank 1 tensors)
An array of numbers is called a vector, or rank 1 tensor, or 1D tensor. A rank 1 tensor is said to have exactly one axis. The following is a tensor vector:
x <- as.array(c(12, 3, 6, 14, 7))
str(x)
num [1:5(1d)] 12 3 6 14 7
length(dim(x))
[1] 1
This vector has five entries and so is called a five-dimensional vector. Don’t confuse a 5D vector with a 5D tensor! A 5D vector has only one axis and has five dimensions along its axis, whereas a 5D tensor has five axes (and may have any number of dimensions along each axis). Dimensionality can denote either the number of entries along a specific axis (as in the case of our 5D vector) or the number of axes in a tensor (such as a 5D tensor), which can be confusing at times. In the latter case, it’s technically more correct to talk about a tensor of rank 5 (the rank of a tensor being the number of axes), but the ambiguous notation 5D tensor is common regardless.
2.2.3 Matrices (rank 2 tensors)
An array of vectors is a matrix, or rank 2 tensor, or 2D tensor. A matrix has two axes (often referred to as rows and columns). You can visually interpret a matrix as a rectangular grid of numbers:
x <- array(seq(3 * 5), dim = c(3, 5))
x

dim(x)
[1] 3 5
The entries from the first axis are called the rows, and the entries from the second axis are called the columns. In the previous example, c(1, 4, 7, 10, 13) is the first row of x, and c(1, 2, 3) is the first column.
2.2.4 Rank 3 and higher-rank tensors
If you supply a length 3 vector to dim, you obtain a rank 3 tensor (or 3D tensor), which you can visually interpret as a cube of numbers or a stack of rank 2 tensors:
x <- array(seq(2 * 3 * 4), dim = c(2, 3, 4))
str(x)
int [1:2, 1:3, 1:4] 1 2 3 4 5 6 7 8 9 10 …
length(dim(x))
[1] 3
By stacking rank 3 tensors, you can create a rank 4 tensor, and so on. In deep learning, you’ll generally manipulate tensors with ranks 0 to 4, although you may go up to 5 if you process video data.
2.2.5 Key attributes
A tensor is defined by the following three key attributes:
- Number of axes (rank)—For instance, a rank 3 tensor has three axes, and a matrix has two axes. This is available from length(dim(x)).
- Shape—This is an integer vector that describes how many dimensions the tensor has along each axis. For instance, the previous matrix example has shape (3, 5), and the rank 3 tensor example has shape (2, 3, 4). A vector has a shape with a single element, such as (5). R arrays don’t distinguish between 1D vectors and scalar tensors, but conceptually, tensors can also be scalar with shape ().
- Data type—This is the type of the data contained in the tensor. R arrays have support for R’s built-in data types like double and integer. Conceptually, however, tensors can support any type of homogeneous data type, and other tensor implementations also provide support for types like like float16, float32, float64 (corresponding to R’s double), int32 (R’s integer type), and so on. In TensorFlow, you are also likely to come across string tensors.
To make this more concrete, let’s look back at the data we processed in the MNIST example. First, we load the MNIST dataset:
library(keras)
mnist <- dataset_mnist()
train_images <- mnist$train$x
train_labels <- mnist$train$y
test_images <- mnist$test$x
test_labels <- mnist$test$y
Next, we display the number of axes of the tensor train_images:
length(dim(train_images))
[1] 3
Here’s its shape:
dim(train_images)
[1] 60000 28 28
And this is its R data type:
typeof(train_images)
[1] "integer"
So what we have here is a rank 3 tensor of integers. More precisely, it’s a stack of 60,000 matrices of 28 × 28 integers. Each such matrix is a grayscale image, with coefficients between 0 and 255 of pixel intensity values.
Let’s display the fifth digit in this rank 3 tensor (see figure 2.2).
Figure 2.2 The fifth sample in our dataset
Listing 2.8 Displaying the fifth digit
digit <- train_images[5, , ]
plot(as.raster(abs(255 - digit), max = 255))
Naturally, the corresponding label is the integer 9:
train_labels[5]
[1] 9
2.2.6 Manipulating tensors in R
In the previous example, we selected a specific digit alongside the first axis using the syntax train_images[i, , ]. Selecting specific elements in a tensor is called tensor slicing. Let’s look at the tensor-slicing operations you can do on R arrays.
NOTE TensorFlow Tensor‘s slicing is similar to R arrays but with some differences. In this section, we focus on R arrays and begin discussing TensorFlow Tensors in chapter 3.
The following example selects digits 10 to 99 and puts them in an array of shape (90, 28, 28):
my_slice <- train_images[10:99, , ]
dim(my_slice)
[1] 90 28 28
In general, you may select slices between any two indices along each tensor axis. For instance, to select 14 × 14 pixels in the bottom-right corner of all images, you would do this:
my_slice <- train_images[, 15:28, 15:28]
dim(my_slice)
[1] 60000 14 14
2.2.7 The notion of data batches
In general, the first axis in all data tensors you’ll come across in deep learning will be the samples axis (sometimes called the samples dimension). In the MNIST example, “samples” are images of digits.
In addition, deep learning models don’t process an entire dataset at once; rather, they break the data into small batches. Concretely, here’s one batch of our MNIST digits, with a batch size of 128:
batch <- train_images[1:128, , ]
And here’s the next batch:
batch <- train_images[129:256, , ]
And the nth batch:
n <- 3
batch <- train_images[seq(to = 128 * n, length.out = 128), , ]
When considering such a batch tensor, the first axis is called the batch axis or batch dimension. This is a term you’ll frequently encounter when using Keras and other deep learning libraries.
2.2.8 Real-world examples of data tensors
Let’s make data tensors more concrete with a few examples similar to what you’ll encounter later. The data you’ll manipulate will almost always fall into one of the following categories:
- Vector data—Rank 2 tensors of shape (samples, features), where each sample is a vector of numerical attributes (“features”)
- Times-series data or sequence data—Rank 3 tensors of shape (samples, timesteps, features), where each sample is a sequence (of length timesteps) of feature vectors
- Images—Rank 4 tensors of shape (samples, height, width, channels), where each sample is a 2D grid of pixels, and each pixel is represented by a vector of values (“channels”)
- Video—Rank 5 tensors of shape (samples, frames, height, width, channels), where each sample is a sequence (of length frames) of images
2.2.9 Vector data
This is one of the most common cases. In such a dataset, each single data point can be encoded as a vector, and thus a batch of data will be encoded as a rank 2 tensor (that is, a matrix), where the first axis is the samples axis and the second axis is the features axis.
Let’s take a look at the next two examples:
- An actuarial dataset of people, where we consider each person’s age, gender, and income. Each person can be characterized as a vector of 3 values, and thus an entire dataset of 100,000 people can be stored in a rank 2 tensor of shape (100000, 3).
- A dataset of text documents, where we represent each document by the counts of how many times each word appears in it (out of a dictionary of 20,000 common words). Each document can be encoded as a vector of 20,000 values (one count per word in the dictionary), and thus an entire dataset of 500 documents can be stored in a tensor of shape (500, 20000).
2.2.10. Time-series data or sequence data
Whenever time matters in your data (or the notion of sequence order), it makes sense to store it in a rank 3 tensor with an explicit time axis. Each sample can be encoded as a sequence of vectors (a rank 2 tensor), and thus a batch of data will be encoded as a rank 3 tensor (see figure 2.3).
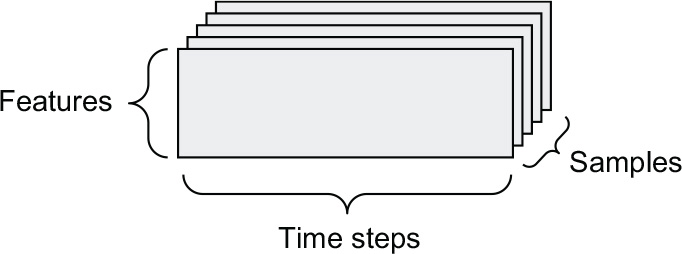
Figure 2.3 A rank 3 time-series data tensor
The time axis is always the second axis by convention. Let’s look at a few examples:
- A dataset of stock prices. Every minute, we store the current price of the stock, the highest price in the past minute, and the lowest price in the past minute. Thus, every minute is encoded as a 3D vector, an entire day of trading is encoded as a matrix of shape (390, 3) (there are 390 minutes in a trading day), and 250 days’ worth of data can be stored in a rank 3 tensor of shape (250, 390, 3). Here, each sample would be one day’s worth of data.
- A dataset of tweets, where we encode each tweet as a sequence of 280 characters out of an alphabet of 128 unique characters. In this setting, each character can be encoded as a binary vector of size 128 (an all-zeros vector except for a 1 entry at the index corresponding to the character). Then each tweet can be encoded as a rank 2 tensor of shape (280, 128), and a dataset of one million tweets can be stored in a tensor of shape (1000000, 280, 128).
2.2.11. Image data
Images typically have three dimensions: height, width, and color depth. Although grayscale images (like our MNIST digits) have only a single color channel and could thus be stored in rank 2 tensors, by convention, image tensors are always rank 3, with a one-dimensional color channel for grayscale images. A batch of 128 grayscale images of size 256 × 256 could thus be stored in a tensor of shape (128, 256, 256, 1), and a batch of 128 color images could be stored in a tensor of shape (128, 256, 256, 3) (see figure 2.4).
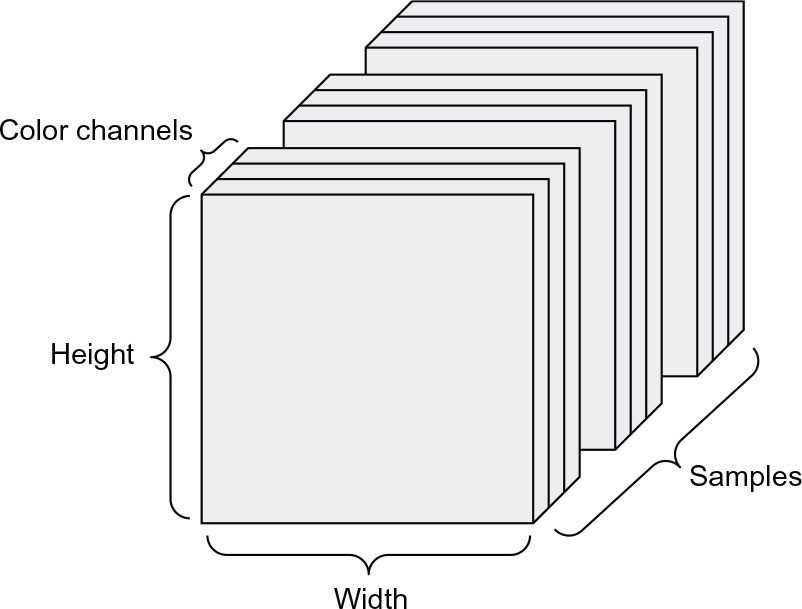
Figure 2.4 A rank 4 image data tensor
Two conventions for shapes of image tensors are the channels-last convention (which is standard in TensorFlow) and the channels-first convention (which is increasingly falling out of favor). The channels-last convention places the color-depth axis at the end: (samples, height, width, color_depth). Meanwhile, the channels-first convention places the color depth axis right after the batch axis: (samples, color_depth, height, width). With the channels-first convention, the previous examples would become (128, 1, 256, 256) and (128, 3, 256, 256). The Keras API provides support for both formats.
2.2.12. Video data
Video data is one of the few types of real-world data for which you’ll need rank 5 tensors. A video can be understood as a sequence of frames, with each frame being a color image. Because each frame can be stored in a rank 3 tensor (height, width, color_depth), a sequence of frames can be stored in a rank 4 tensor (frames, height, width, color_depth), and thus a batch of different videos can be stored in a rank 5 tensor of shape (samples, frames, height, width, color_depth).
For instance, a 60-second, 144 × 256 YouTube video clip sampled at 4 frames per second would have 240 frames. A batch of four such video clips would be stored in a tensor of shape (4, 240, 144, 256, 3). That’s a total of 106,168,320 values! If the data type of the tensor was R integers, each value would be stored in 32 bits, so the tensor would represent 405 MB. Heavy! Videos you encounter in real life are much lighter, because they aren’t stored as R integers, and they’re typically compressed by a large factor (such as in the MPEG format).
2.3 The gears of neural networks: Tensor operations
As much as any computer program can be ultimately reduced to a small set of binary operations on binary inputs (AND, OR, NOR, and so on), all transformations learned by deep neural networks can be reduced to a handful of tensor operations (or tensor functions) applied to tensors of numeric data. For instance, it’s possible to add tensors, multiply tensors, and so on. In our initial example, we built our model by stacking Dense layers on top of each other. A Keras layer instance looks like this:
layer_dense(units = 512, activation = "relu")
<keras.layers.core.dense.Dense object at 0x7f7b0e8cf520>
This layer can be interpreted as a function, which takes as input a matrix and returns another matrix—a new representation for the input tensor. Specifically, the function is as follows (where W is a matrix and b is a vector, both properties of the layer):
output <- relu(dot(W, input) + b)
Let’s unpack this. We have the following three tensor operations here:
- A dot product (dot) between the input tensor and a tensor named W
- An addition (+) between the resulting matrix and a vector b
- A relu operation: relu(x) is an element-wise max(x, 0); relu stands for rectified linear unit
Although this section deals entirely with linear algebra expressions, you won’t find any mathematical notation here. I’ve found that mathematical concepts can be more readily mastered by programmers with no mathematical background if they’re expressed as short code snippets instead of mathematical equations. So we’ll use R and TensorFlow code throughout.
2.3.1 Element-wise operations
The relu operation and addition are element-wise operations: operations that are applied independently to each entry in the tensors being considered. This means these operations are highly amenable to massively parallel implementations (vectorized implementations, a term that comes from the vector processor supercomputer architecture from the 1970–1990 period). If you want to write a naive R implementation of an element-wise operation, you use a for loop, as in the following naive implementation of an element-wise relu operation:
naive_relu <- functsion(x) {
stopifnot(length(dim(x)) == 2)➊
for (i in 1:nrow(x))
for (j in 1:ncol(x))
x[i, j] <- max(x[i, j], 0)
x
}
➊ x is a rank 2 tensor (a matrix).
You could do the same for addition:
naive_add <- function(x, y) {
stopifnot(length(dim(x)) == 2, dim(x) == dim(y))➊
for (i in 1:nrow(x))
for (j in 1:ncol(x))
x[i, j] <- x[i, j] + y[i, j]
x
}
➊ x and y are rank 2 tensors.
On the same principle, you can do element-wise multiplication, subtraction, and so on.
In practice, when dealing with R arrays, these operations are available as well-optimized built-in R functions, which themselves delegate the heavy lifting to a Basic Linear Algebra Subprograms (BLAS) implementation. BLAS are low-level, highly parallel, efficient tensor-manipulation routines that are typically implemented in Fortran or C. So, in R, you can do the following element-wise operation, and it will be blazing fast:
z <- x + y➊
z[z < 0] <- 0➋
➊ Element-wise addition
➋ Element-wise relu
Let’s actually time the difference here:
random_array <- function(dim, min = 0, max = 1)
array(runif(prod(dim), min, max),
dim)
x <- random_array(c(20, 100))
y <- random_array(c(20, 100))
system.time({
for (i in seq_len(1000)) {
z <- x + y
z[z < 0] <- 0
}
})[["elapsed"]]
[1] 0.009
This takes 0.009 seconds. Meanwhile, the naive version takes a stunning 0.72 seconds:
system.time({
for (i in seq_len(1000)) {
z <- naive_add(x, y)
z <- naive_relu(z)
}
})[["elapsed"]]
[1] 0.724
Likewise, when running TensorFlow code on a GPU, element-wise operations are executed via fully vectorized CUDA implementations that can best utilize the highly parallel GPU chip architecture.
2.3.2 Broadcasting
Our earlier naive implementation of naive_add supports only the addition of rank 2 tensors with identical shapes. But in the layer_dense() introduced earlier, we added a rank 2 tensor with a vector. What happens with addition when the shapes of the two tensors being added differ?
What we’d like is for the smaller tensor to be broadcast to match the shape of the larger tensor. Broadcasting consists of the following two steps:
- 1 Axes (called broadcast axes) are added to the smaller tensor to match the length(dim(x)) of the larger tensor.
- 2 The smaller tensor is repeated alongside these new axes to match the full shape of the larger tensor.
Note that Tensorflow Tensors, covered in chapter 3, have rich broadcasting functionality built in. Here, however, we are building up machine learning concepts from scratch using R arrays and are intentionally avoiding R’s implicit recycling behavior when operating on two arrays of different dimensions. We can implement our own recycling approach by building up the smaller tensor to match the shape of the larger tensor, at which point we are again back to doing a standard element-wise operation.
Let’s look at a concrete example. Consider X with shape (32, 10) and y with shape (10):
X <- random_array(c(32, 10))➊
y <- random_array(c(10))➋
➊ X is a random matrix with shape (32, 10).
➋ y is a random vector with shape (10).
First, we add a size 1 first axis to y, whose shape becomes (1, 10):
dim(y) <- c(1, 10)
str(y)➊
num [1, 1:10] 0.885 0.429 0.737 0.553 0.426 …
➊ The shape of y is now (1, 10).
Then, we repeat y 32 times alongside this new axis, so that we end up with a tensor Y with shape (32, 10), where Y[i, ] == y for i in seq(32):
Y <- y[rep(1, 32), ]➊
str(Y)
num [1:32, 1:10] 0.885 0.885 0.885 0.885 0.885 …
➊ Repeat y 32 times along axis 1 to obtain Y, which has shape (32, 10).
At this point, we can proceed to add X and Y, because they have the same shape.
In terms of implementation, ideally we want no new rank 2 tensor to be created, because that is terribly inefficient. In most tensor implementations, including R and TensorFlow, the repetition operation is entirely virtual: it happens at the algorithmic level rather than at the memory level. However, be aware that R’s recycling and TensorFlow’s (and NumPy’s) broadcasting differ in their behavior (we go into details in chapter 3). Regardless, thinking of the vector being repeated 10 times alongside a new axis is a helpful mental model. Here’s what a naive implementation would look like:
naive_add_matrix_and_vector <- function(x, y) {
stopifnot(length(dim(x)) == 2,➊
length(dim(y)) == 1,➋
ncol(x) == dim(y))
for (i in seq(dim(x)[1]))
for (j in seq(dim(x)[2]))
x[i, j] <- x[i, j] + y[j]
x
}
➊ x is a rank 2 tensor.
➋ y is a vector.
2.3.3 Tensor product
The tensor product, or dot product (not to be confused with an element-wise product, the * operator), is one of the most common, most useful tensor operations. In R, an element-wise product is done with the * operator, whereas dot products use the %*% operator:
x <- random_array(c(32))
y <- random_array(c(32))
z <- x %*% y
In mathematical notation, you’d note the operation with a dot (•):
z = x • y
Mathematically, what does the dot operation do? Let’s start with the dot product of two vectors, x and y. It’s computed as follows:
naive_vector_dot <- function(x, y) {
stopifnot(length(dim(x)) == 1,➊
length(dim(y)) == 1,➊
dim(x) == dim(y))
z <- 0
for (i in seq_along(x))
z <- z + x[i] * y[i]
z
}
➊ x and y are 1D vectors of the same size.
You’ll have noticed that the dot product between two vectors is a scalar and that only vectors with the same number of elements are compatible for a dot product.
You can also take the dot product between a matrix x and a vector y, which returns a vector where the coefficients are the dot products between y and the rows of x:
naive_matrix_vector_dot <- function(x, y) {
stopifnot(length(dim(x)) == 2,➊
length(dim(y)) == 1,➋
nrow(x) == dim(y))➌
z <- array(0, dim = dim(y))➍
for (i in 1:nrow(x))
for (j in 1:ncol(x))
z[i] <- z[i] + x[i, j] * y[j]
z
}
➊ x is a 2D tensor (matrix).
➋ y is a 1D tensor (vector).
➌ The first dimension of x must be the same as the first dimension of y!
➍ This operation returns a vector of zeros with the same shape as y.
You could also reuse the code we wrote previously, which highlights the relationship between a matrix-vector product and a vector product:
naive_matrix_vector_dot <- function(x, y) {
z <- array(0, dim = c(nrow(x)))
for (i in 1:nrow(x))
z[i] <- naive_vector_dot(x[i, ], y)
z
}
Note that as soon as one of the two tensors has a length(dim(x)) greater than 1, %*% is no longer symmetric, which is to say that x %*% y isn’t the same as y %*% x.
Of course, a dot product generalizes to tensors with an arbitrary number of axes. The most common applications may be the dot product between two matrices. You can take the dot product of two matrices x and y (x %*% y) if and only if ncol(x) == nrow(y). The result is a matrix with shape (nrow(x), ncol(y)), where the coefficients are the vector products between the rows of x and the columns of y. The naive implementation is shown here:
naive_matrix_dot <- function(x, y) {
stopifnot(length(dim(x)) == 2,➊
length(dim(y)) == 2,
ncol(x) == nrow(y))➋
z <- array(0, dim = c(nrow(x), ncol(y)))➌
for (i in 1:nrow(x))➍
for (j in 1:ncol(y)) {➎
row_x <- x[i, ]
column_y <- y[, j]
z[i, j] <- naive_vector_dot(row_x, column_y)
}
z
}
➊ x and y are 2D tensors (matrices).
➋ The first dimension of x must be the same as the first dimension of y!
➌ This operation returns a matrix of zeros with a specific shape.
➍ Iterate over the rows of x…
➎ … and over the columns of y.
To understand dot-product shape compatibility, it helps to visualize the input and output tensors by aligning them as shown in figure 2.5.
In the figure, x, y, and z are pictured as rectangles (literal boxes of coefficients). Because the rows of x and the columns of y must have the same size, it follows that the width of x must match the height of y. If you go on to develop new machine learning algorithms, you’ll likely be drawing such diagrams often.
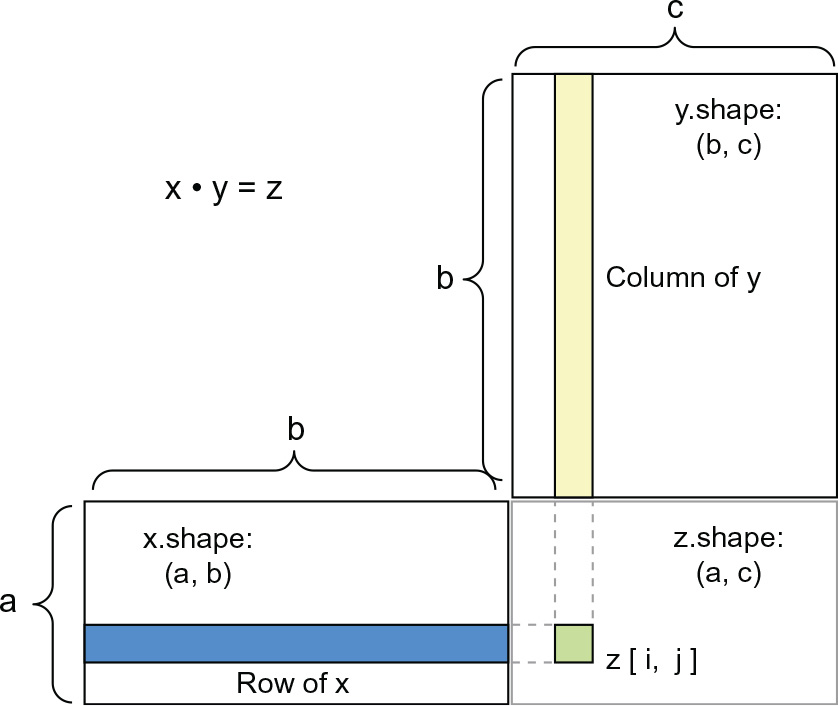
Figure 2.5 Matrix dot-product box diagram
More generally, you can take the dot product between higher-dimensional tensors, following the same rules for shape compatibility as outlined earlier for the 2D case:
(a, b, c, d) • (d) -> (a, b, c)
(a, b, c, d) • (d, e) -> (a, b, c, e)
And so on.
2.3.4 Tensor reshaping
A third type of tensor operation that’s essential to understand is tensor reshaping. Although it wasn’t used in the layer_dense() in our first neural network example, we used it when we preprocessed the digits data before feeding it into our model, as shown next:
train_images <- array_reshape(train_images, c(60000, 28 * 28))
Note that we use the array_reshape() function rather than the `dim<-`() function to reshape R arrays. This is so that the data is reinterpreted using row-major semantics (as opposed to R’s default column-major semantics), which is in turn compatible with the way the numerical libraries called by Keras (NumPy, TensorFlow, and so on) interpret array dimensions. You should always use the array_reshape() function when reshaping R arrays that will be passed to Keras.
Reshaping a tensor means rearranging its rows and columns to match a target shape. Naturally, the reshaped tensor has the same total number of coefficients as the initial tensor. Reshaping is best understood via simple examples:
x <- array(1:6)
x
[1] 1 2 3 4 5 6
array_reshape(x, dim = c(3, 2))
[,1] [,2]
[1,] 1 2
[2,] 3 4
[3,] 5 6
array_reshape(x, dim = c(2, 3))
[,1] [,2] [,3]
[1,] 1 2 3
[2,] 4 5 6
A special case of reshaping that’s commonly encountered is transposition. Transposing a matrix means exchanging its rows and its columns, so that x[i, ] becomes x[, i]. We can use the t() function to transpose a matrix:
x <- array(1:6, dim = c(3, 2))
x
[,1] [,2]
[1,] 1 4
[2,] 2 5
[3,] 3 6
t(x)
[,1] [,2] [,3]
[1,] 1 2 3
[2,] 4 5 6
2.3.5 Geometric interpretation of tensor operations
Because the contents of the tensors manipulated by tensor operations can be interpreted as coordinates of points in some geometric space, all tensor operations have a geometric interpretation. For instance, let’s consider addition. We’ll start with the following vector:
A = c(0.5, 1)
It’s a point in a 2D space (see figure 2.6). It’s common to picture a vector as an arrow linking the origin to the point, as shown in figure 2.7.
Let’s consider a new point, B = c(1, 0.25), which we’ll add to the previous one. This is done geometrically by chaining together the vector arrows, with the resulting location being the vector representing the sum of the previous two vectors (see figure 2.8). As you can see, adding a vector B to a vector A represents the action of copying point A in a new location, whose distance and direction from the original point A is determined by the vector B. If you apply the same vector addition to a group of points in the plane (an object), you would be creating a copy of the entire object in a new location (see figure 2.9). Tensor addition thus represents the action of translating an object (moving the object without distorting it) by a certain amount in a certain direction.
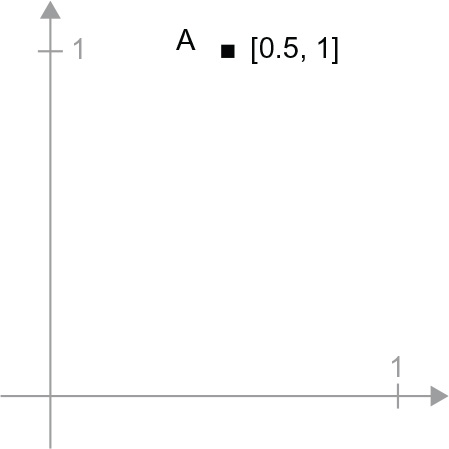
Figure 2.6 A point in a 2D space
Figure 2.7 A point in a 2D space pictured as an arrow
In general, elementary geometric operations such as translation, rotation, scaling, skewing, and so on can be expressed as tensor operations. Here are a few examples:
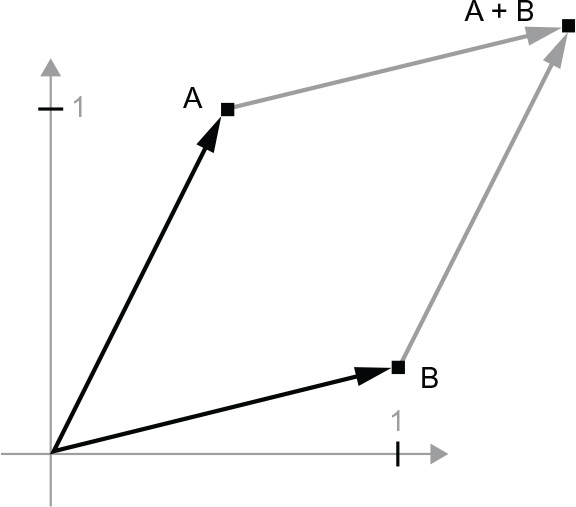
Figure 2.8 Geometric interpretation of the sum of two vectors
- Translation—As you just saw, adding a vector to a point will move the point by a fixed amount in a fixed direction. Applied to a set of points (such as a 2D object), this is called a “translation” (see figure 2.9).

Figure 2.9 2D translation as a vector addition
- Rotation—A counterclockwise rotation of a 2D vector by an angle theta (see figure 2.10) can be achieved via a dot product with a 2 × 2 matrix R = rbind(c(cos(theta), -sin(theta)), c(sin(theta), cos(theta)).

Figure 2.10 2D rotation (counterclock-wise) as a dot product
- Scaling—A vertical and horizontal scaling of the image (see figure 2.11) can be achieved via a dot product with a 2 × 2 matrix S = rbind(c(horizontal_factor, 0), c(0, vertical_factor)) (note that such a matrix is called a diagonal matrix, because it has only non-zero coefficients in its “diagonal,” going from the top left to the bottom right).
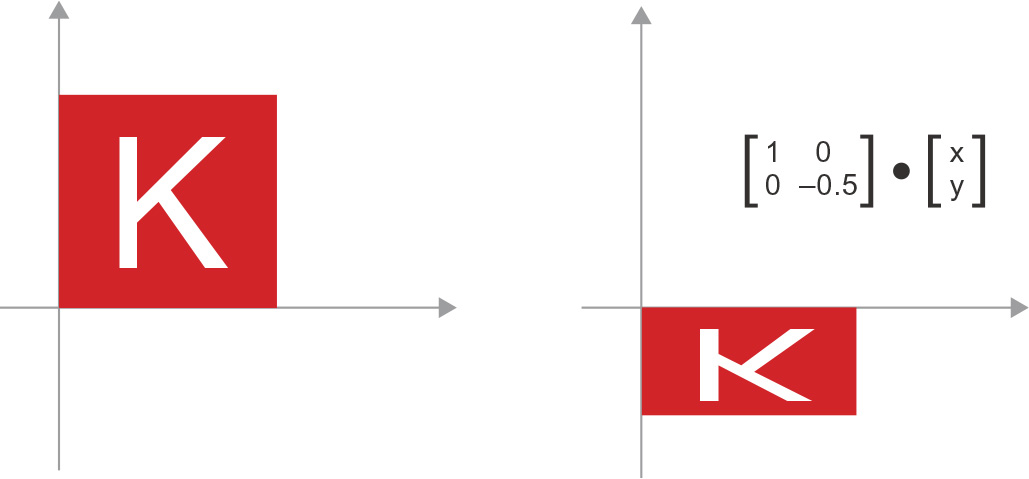
Figure 2.11 2D scaling as a dot product
- Linear transform—A dot product with an arbitrary matrix implements a linear transform. Note that scaling and rotation, listed previously, are by definition linear transforms.
- Affine transform—An affine transform (see figure 2.12) is the combination of a linear transform (achieved via a dot product with some matrix) and a translation (achieved via a vector addition). As you have probably recognized, that’s exactly the y = W • x + b computation implemented by layer_dense()! A Dense layer without an activation function is an affine layer.

Figure 2.12 Affine transform in the plane
- Dense layer with relu activation—An important observation about affine transforms is that if you apply many of them repeatedly, you still end up with an affine transform (so you could just have applied that one affine transform in the first place). Let’s try it with two: affine2(affine1(x)) = W2 • (W1 • x + b1) + b2 = (W2 • W1) • x + (W2 • b1 + b2). That’s an affine transform where the linear part is the matrix W2 • W1 and the translation part is the vector W2 • b1 + b2. As a consequence, a multilayer neural network made entirely of Dense layers without activations would be equivalent to a single Dense layer. This “deep” neural network would just be a linear model in disguise! This is why we need activation functions, like relu (seen in action in figure 2.13). Thanks to activation functions, a chain of Dense layers can be made to implement very complex, nonlinear geometric transformations, resulting in very rich hypothesis spaces for your deep neural networks. We’ll cover this idea in more detail in the next chapter.

Figure 2.13 Affine transform followed by relu activation
2.3.6 A geometric interpretation of deep learning
You just learned that neural networks consist entirely of chains of tensor operations, and that these tensor operations are just simple geometric transformations of the input data. It follows that you can interpret a neural network as a very complex geometric transformation in a high-dimensional space, implemented via a series of simple steps.
In 3D, the following mental image may prove useful. Imagine two sheets of colored paper: one red and one blue. Put one on top of the other. Now crumple them together into a small ball. That crumpled paper ball is your input data, and each sheet of paper is a class of data in a classification problem. What a neural network is meant to do is figure out a transformation of the paper ball that would uncrumple it, so as to make the two classes cleanly separable again (see figure 2.14). With deep learning, this would be implemented as a series of simple transformations of the 3D space, such as those you could apply on the paper ball with your fingers, one movement at a time.

Figure 2.14 Uncrumpling a complicated manifold of data
Uncrumpling paper balls is what machine learning is about: finding neat representations for complex, highly folded data manifolds in high-dimensional spaces (a manifold is a continuous surface, like our crumpled sheet of paper). At this point, you should have a pretty good intuition as to why deep learning excels at this: it takes the approach of incrementally decomposing a complicated geometric transformation into a long chain of elementary ones, which is pretty much the strategy a human would follow to uncrumple a paper ball. Each layer in a deep network applies a transformation that disentangles the data a little, and a deep stack of layers makes tractable an extremely complicated disentanglement process.
2.4 The engine of neural networks: Gradient-based optimization
As you saw in the previous section, each neural layer from our first model example transforms its input data as follows:
output <- relu(dot(input, W) + b)
In this expression, W and b are tensors that are attributes of the layer. They’re called the weights or trainable parameters of the layer (the kernel and bias attributes, respectively). These weights contain the information learned by the model from exposure to training data.
Initially, these weight matrices are filled with small random values (a step called random initialization). Of course, there’s no reason to expect that relu(dot(input, W) + b), when W and b are random, will yield any useful representations. The resulting representations are meaningless—but they’re a starting point. What comes next is to gradually adjust these weights, based on a feedback signal. This gradual adjustment, also called training, is the learning that machine learning is all about. This happens within what’s called a training loop, which works as follows. Repeat these steps in a loop, until the loss seems sufficiently low:
- 1 Draw a batch of training samples, x, and corresponding targets, y_true.
- 2 Run the model on x (a step called the forward pass) to obtain predictions, y_pred.
- 3 Compute the loss of the model on the batch, a measure of the mismatch between y_pred and y_true.
- 4 Update all weights of the model in a way that slightly reduces the loss on this batch.
You’ll eventually end up with a model that has a very low loss on its training data: a low mismatch between predictions, y_pred, and expected targets, y_true. The model has “learned” to map its inputs to correct targets. From afar, it may look like magic, but when you reduce it to elementary steps, it turns out to be simple.
Step 1 sounds easy enough—just I/O code. Steps 2 and 3 are merely the application of a handful of tensor operations, so you could implement these steps purely from what you learned in the previous section. The difficult part is step 4: updating the model’s weights. Given an individual weight coefficient in the model, how can you compute whether the coefficient should be increased or decreased, and by how much?
One naive solution would be to freeze all weights in the model except the one scalar coefficient being considered, and try different values for this coefficient. Let’s say the initial value of the coefficient is 0.3. After the forward pass on a batch of data, the loss of the model on the batch is 0.5. If you change the coefficient’s value to 0.35 and rerun the forward pass, the loss increases to 0.6. But if you lower the coefficient to 0.25, the loss falls to 0.4. In this case, it seems that updating the coefficient by –0.05 would contribute to minimizing the loss. This would have to be repeated for all coefficients in the model.
But such an approach would be horribly inefficient, because you’d need to compute two forward passes (which are expensive) for every individual coefficient (of which there are many—usually thousands and sometimes up to millions). Thankfully, there’s a much better approach: gradient descent.
Gradient descent is the optimization technique that powers modern neural networks. Here’s the gist of it: all of the functions used in our models (such as dot or +) transform their input in a smooth and continuous way. If you look at z = x + y, for instance, a small change in y results in only a small change in z, and if you know the direction of the change in y, you can infer the direction of the change in z. Mathematically, you’d say these functions are differentiable. If you chain together such functions, the bigger function you obtain is still differentiable. In particular, this applies to the function that maps the model’s coefficients to the loss of the model on a batch of data: a small change in the model’s coefficients results in a small, predictable change in the loss value. This enables you to use a mathematical operator called the gradient to describe how the loss varies as you move the model’s coefficients in different directions. If you compute this gradient, you can use it to move the coefficients (all at once in a single update, rather than one at a time) in a direction that decreases the loss.
If you already know what differentiable means and what a gradient is, you can skip to section 2.4.3. Otherwise, the following two sections will help you understand these concepts.
2.4.1 What’s a derivative?
Consider a continuous, smooth function f(x) = y, mapping a number, x, to a new number, y. We can use the function in figure 2.15 as an example.
Because the function is continuous, a small change in x can only result in a small change in y—that’s the intuition behind continuity. Let’s say you increase x by a small factor, epsilon_x: this results in a small epsilon_y change to y, as shown in figure 2.16.
In addition, because the function is smooth (its curve doesn’t have any abrupt angles), when epsilon_x is small enough, around a certain point p, it’s possible to approximate f as a linear function of slope a, so that epsilon_y becomes a * epsilon_x:
f(x + epsilon_x) = y + a * epsilon_x
Obviously, this linear approximation is valid only when x is close enough to p.
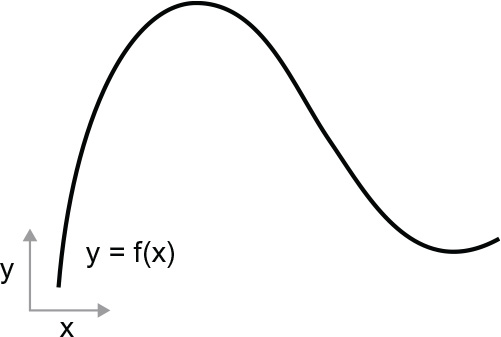
Figure 2.15 A continuous, smooth function
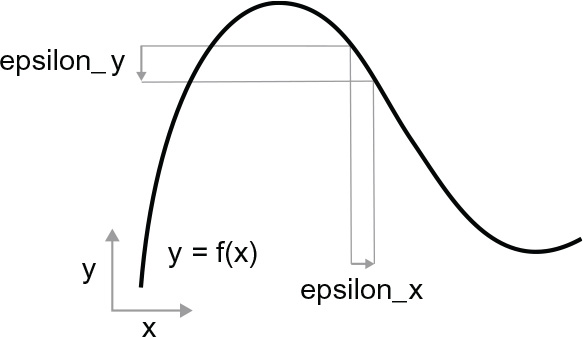
Figure 2.16 With a continuous function, a small change in x results in a small change in y.

Figure 2.17 Derivative of f in p
The slope a is called the derivative of f in p. If a is negative, it means a small increase in x around p will result in a decrease of f(x) (as shown in figure 2.17), and if a is positive, a small increase in x will result in an increase of f(x). Further, the absolute value of a (the magnitude of the derivative) tells you how quickly this increase or decrease will happen.
For every differentiable function f(x) (differentiable means “can be differentiated to find the derivative”: for example, smooth, continuous functions can be differentiated), there exists a derivative function f’(x) that maps values of x to the slope of the local linear approximation of f in those points. For instance, the derivative of cos(x) is -sin(x), the derivative of f(x) = a * x is f’(x) = a, and so on.
Being able to differentiate functions is a very powerful tool when it comes to optimization, the task of finding values of x that minimize the value of f(x). If you’re trying to update x by a factor epsilon_x to minimize f(x), and you know the derivative of f, then your job is done: the derivative completely describes how f(x) evolves as you change x. If you want to reduce the value of f(x), you just need to move x a little in the opposite direction from the derivative.
2.4.2 Derivative of a tensor operation: The gradient
The function we were just looking at turned a scalar value x into another scalar value y: you could plot it as a curve in a 2D plane. Now imagine a function that turns a list of scalars (x, y) into a scalar value z: that would be a vector operation. You could plot it as a 2D surface in a 3D space (indexed by coordinates x, y, z). Likewise, you can imagine functions that take matrices as inputs, functions that take rank 3 tensors as inputs, and so on.
The concept of differentiation can be applied to any such function, as long as the surfaces they describe are continuous and smooth. The derivative of a tensor operation (or tensor function) is called a gradient. Gradients are just the generalization of the concept of derivatives to functions that take tensors as inputs. Remember how, for a scalar function, the derivative represents the local slope of the curve of the function? In the same way, the gradient of a tensor function represents the curvature of the multidimensional surface described by the function. It characterizes how the output of the function varies when its input parameters vary.
Let’s look at an example grounded in machine learning. Consider the following:
- An input vector, x (a sample in a dataset)
- A matrix, W (the weights of a model)
- A target, y_true (what the model should learn to associate to x)
- A loss function, loss_fn() (meant to measure the gap between the model’s current predictions and y_true)
You can use W to compute a target candidate y_pred, and then compute the loss, or mismatch, between the target candidate y_pred and the target y_true:
y_pred <- dot(W, x)➊
loss_value <- loss_fn(y_pred, y_true)➋
➊ We use the model weights, W, to make a prediction for x.
➋ We estimate how far off the prediction was.
Now we’d like to use gradients to figure out how to update W so as to make loss_value smaller. How do we do that? Given fixed inputs x and y_true, the preceding operations can be interpreted as a function mapping values of W (the model’s weights) to loss values:
loss_value <- f(W)➊
➊ f() describes the curve (or high-dimensional surface) formed by loss values when W varies.
Let’s say the current value of W is W0. Then the derivative of f() at the point W0 is a tensor grad(loss_value, W0), with the same shape as W, where each coefficient grad(loss_ value, W0)[i, j] indicates the direction and magnitude of the change in loss_value you observe when modifying W0[i, j]. That tensor grad(loss_value, W0) is the gradient of the function f(W) = loss_value in W0, also called “gradient of loss_value with respect to W around W0.”
Partial derivatives
The tensor operation grad(f(W), W) (which takes as input a matrix W) can be expressed as a combination of scalar functions, grad_ij(f(W), w_ij), each of which would return the derivative of loss_value = f(W) with respect to the coefficient W[i, j] of W, assuming all other coefficients are constant. grad_ij is called the partial derivative of f with respect to W[i, j].
Concretely, what does grad(loss_value, W0) represent? You saw earlier that the derivative of a function f(x) of a single coefficient can be interpreted as the slope of the curve of f(). Likewise, grad(loss_value, W0) can be interpreted as the tensor describing the direction of steepest ascent of loss_value = f(W) around W0, as well as the slope of this ascent. Each partial derivative describes the slope of f() in a specific direction.
For this reason, in much the same way that, for a function f(x), you can reduce the value of f(x) by moving x a little in the opposite direction from the derivative, with a function f(W) of a tensor, you can reduce loss_value = f(W) by moving W in the opposite direction from the gradient: for example, W1 = W0 – step * grad(f(W0), W0) (where step is a small scaling factor). That means going against the direction of steepest ascent of f, which intuitively should put you lower on the curve. Note that the scaling factor step is needed because grad(loss_value, W0) approximates the curvature only when you’re close to W0, so you don’t want to get too far from W0.
2.4.3 Stochastic gradient descent
Given a differentiable function, it’s theoretically possible to find its minimum analytically: it’s known that a function’s minimum is a point where the derivative is 0, so all you have to do is find all the points where the derivative goes to 0 and check for which of these points the function has the lowest value.
Applied to a neural network, that means finding analytically the combination of weight values that yields the smallest possible loss function. This can be done by solving the equation grad(f(W), W) = 0 for W. This is a polynomial equation of N variables, where N is the number of coefficients in the model. Although it would be possible to solve such an equation for N = 2 or N = 3, doing so is intractable for real neural networks, where the number of parameters is never less than a few thousand and can often be several tens of millions.
Instead, you can use the four-step algorithm outlined at the beginning of this section: modify the parameters little by little based on the current loss value for a random batch of data, as follows. Because you’re dealing with a differentiable function, you can compute its gradient, which gives you an efficient way to implement step 4. If you update the weights in the opposite direction from the gradient, the loss will be a little less every time:
- 1 Draw a batch of training samples, x, and corresponding targets, y_true.
- 2 Run the model on x to obtain predictions, y_pred (this is called the forward pass).
- 3 Compute the loss of the model on the batch, a measure of the mismatch between y_pred and y_true.
- 4 Compute the gradient of the loss with regard to the model’s parameters (this is called the backward pass).
- 5 Move the parameters a little in the opposite direction from the gradient—for example, W = W – (learning_rate * gradient)—thus reducing the loss on the batch a bit. The learning rate (learning_rate here) would be a scalar factor modulating the “speed” of the gradient descent process.
Easy enough! What we just described is called mini-batch stochastic gradient descent (mini-batch SGD). The term stochastic refers to the fact that each batch of data is drawn at random (stochastic is a scientific synonym of random). Figure 2.18 illustrates what happens in 1D, when the model has only one parameter and you have only one training sample.
As you can see, intuitively it’s important to pick a reasonable value for the learning_ rate factor. If it’s too small, the descent down the curve will take many iterations, and it could get stuck in a local minimum. If learning_rate is too large, your updates may end up taking you to completely random locations on the curve.
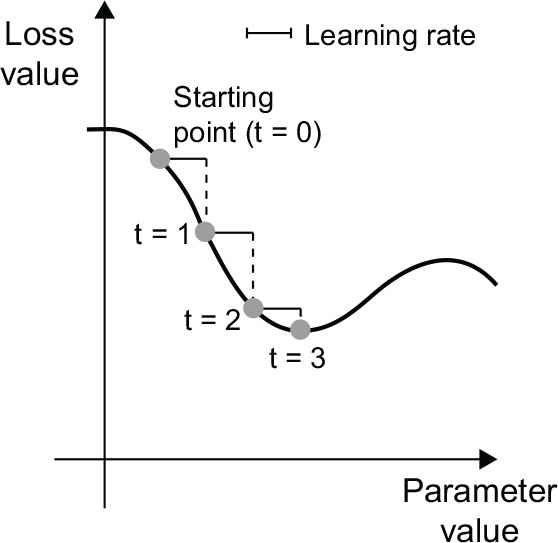
Figure 2.18 SGD down a 1D loss curve (one learnable parameter)
Note that a variant of the mini-batch SGD algorithm would be to draw a single sample and target at each iteration, rather than drawing a batch of data. This would be true SGD (as opposed to mini-batch SGD). Alternatively, going to the opposite extreme, you could run every step on all data available, which is called batch gradient descent. Each update would then be more accurate but far more expensive. The efficient compromise between these two extremes is to use mini-batches of reasonable size.
Although figure 2.18 illustrates gradient descent in a 1D parameter space, in practice you’ll use gradient descent in highly dimensional spaces: every weight coefficient in a neural network is a free dimension in the space, and there may be tens of thousands or even millions of them. To help you build intuition about loss surfaces, you can also visualize gradient descent along a 2D loss surface, as shown in figure 2.19. But you can’t possibly visualize what the actual process of training a neural network looks like—you can’t represent a 1,000,000-dimensional space in a way that makes sense to humans. As such, it’s good to keep in mind that the intuitions you develop through these low-dimensional representations may not always be accurate in practice. This has historically been a source of issues in the world of deep learning research.

Figure 2.19 Gradient descent down a 2D loss surface (two learnable parameters)
Additionally, multiple variants of SGD exist that differ by taking into account previous weight updates when computing the next weight update, rather than just looking at the current value of the gradients. There is, for instance, SGD with momentum, as well as AdaGrad, RMSprop, and several others. Such variants are known as optimization methods or optimizers. In particular, the concept of momentum, which is used in many of these variants, deserves your attention. Momentum addresses two issues with SGD: convergence speed and local minima. Consider figure 2.20, which shows the curve of a loss as a function of a model parameter.

Figure 2.20 A local minimum and a global minimum
As you can see, around a certain parameter value, there is a local minimum: around that point, moving left would result in the loss increasing, but so would moving right. If the parameter under consideration were being optimized via SGD with a small learning rate, the optimization process could get stuck at the local minimum instead of making its way to the global minimum.
You can avoid such issues by using momentum, which draws inspiration from physics. A useful mental image here is to think of the optimization process as a small ball rolling down the loss curve. If it has enough momentum, the ball won’t get stuck in a ravine and will end up at the global minimum. Momentum is implemented by moving the ball at each step based not only on the current slope value (current acceleration) but also on the current velocity (resulting from past acceleration). In practice, this means updating the parameter w based not only on the current gradient value but also on the previous parameter update, such as in this naive implementation:
past_velocity <- 0
momentum <- 0.1➊
repeat {➋
p <- get_current_parameters()➌
if (p$loss <= 0.01)
break
velocity <- past_velocity * momentum + learning_rate * p$gradient w <- p$w + momentum * velocity - learning_rate * p$gradient
past_velocity <- velocity
update_parameter(w)
}
➊ Constant momentum factor
➋ Optimization loop
➌ p contains: w, loss, gradient
2.4.4 Chaining derivatives: The backpropagation algorithm
In the preceding algorithm, we casually assumed that because a function is differentiable, we can easily compute its gradient. But is that true? How can we compute the gradient of complex expressions in practice? In the two-layer model we started the chapter with, how can we get the gradient of the loss with regard to the weights? That’s where the backpropagation algorithm comes in.
THE CHAIN RULE
Backpropagation is a way to use the derivatives of simple operations (such as addition, relu, or tensor product) to easily compute the gradient of arbitrarily complex combinations of these atomic operations. Crucially, a neural network consists of many tensor operations chained together, each of which has a simple, known derivative. For instance, the model defined in listing 2.2 can be expressed as a function parameterized by the variables W1, b1, W2, and b2 (belonging to the first and second Dense layers, respectively), involving the atomic operations dot, relu, softmax, and +, as well as our loss function loss, which are all easily differentiable:
loss_value <- loss(y_true,
softmax(dot(relu(dot(inputs, W1) + b1), W2) + b2))
Calculus tells us that such a chain of functions can be derived using the following identity, called the chain rule.
Consider two functions f and g, as well as the composed function fg such that fg(x) == f(g(x)):
fg <- function(x) {
x1 <- g(x)
y <- f(x1)
y
}
Then the chain rule states that grad(y, x) == grad(y, x1) * grad(x1, x). This enables you to compute the derivative of fg as long as you know the derivatives of f and g. The chain rule is named as it is because when you add more intermediate functions, it starts looking like a chain:
fghj <- function(x) {
x1 <- j(x)
x2 <- h(x1)
x3 <- g(x2)
y <- f(x3)
y
}
grad(y, x) == (grad(y, x3) * grad(x3, x2) *
![]() grad(x2, x1) * grad(x1, x))
grad(x2, x1) * grad(x1, x))
Applying the chain rule to the computation of the gradient values of a neural network gives rise to an algorithm called backpropagation. Let’s see how that works, concretely.
AUTOMATIC DIFFERENTIATION WITH COMPUTATION GRAPHS
A useful way to think about backpropagation is in terms of computation graphs. A computation graph is the data structure at the heart of TensorFlow and the deep learning revolution in general. It’s a directed acyclic graph of operations—in our case, tensor operations. For instance, figure 2.21 shows the graph representation of our first model.
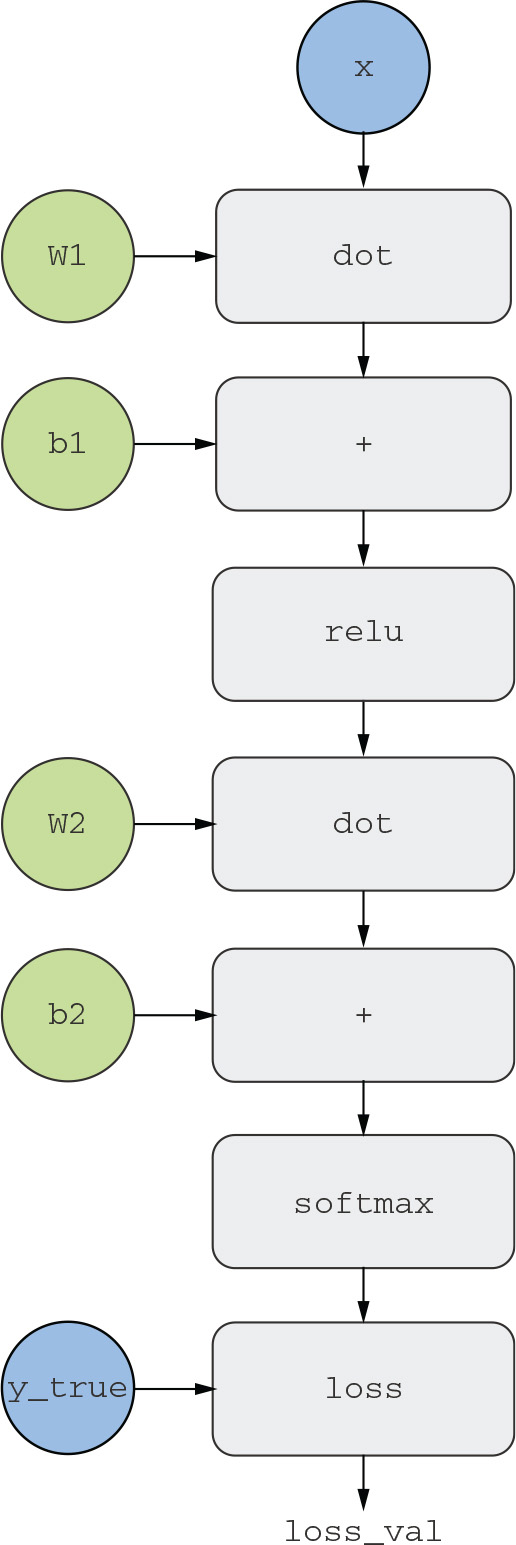
Figure 2.21 The computation graph representation of our two-layer model
Computation graphs have been an extremely successful abstraction in computer science because they enable us to treat computation as data: a computable expression is encoded as a machine-readable data structure that can be used as the input or output of another program. For instance, you could imagine a program that receives a computation graph and returns a new computation graph that implements a large-scale distributed version of the same computation. This would mean that you could distribute any computation without having to write the distribution logic yourself. Or imagine a program that receives a computation graph and can automatically generate the derivative of the expression it represents. It’s much easier to do these things if your computation is expressed as an explicit graph data structure rather than, say, lines of ASCII characters in a .R file.
To explain backpropagation clearly, let’s look at a really basic example of a computation graph (see figure 2.22). We’ll consider a simplified version of figure 2.21, where we have only one linear layer and where all variables are scalar. We’ll take two scalar variables w and b, a scalar input x, and apply some operations to them to combine them into an output y. Finally, we’ll apply an absolute value error-loss function: loss_val = abs(y_true - y). Because we want to update w and b in a way that will minimize loss_val, we are interested in computing grad(loss_val, b) and grad(loss_val, w).
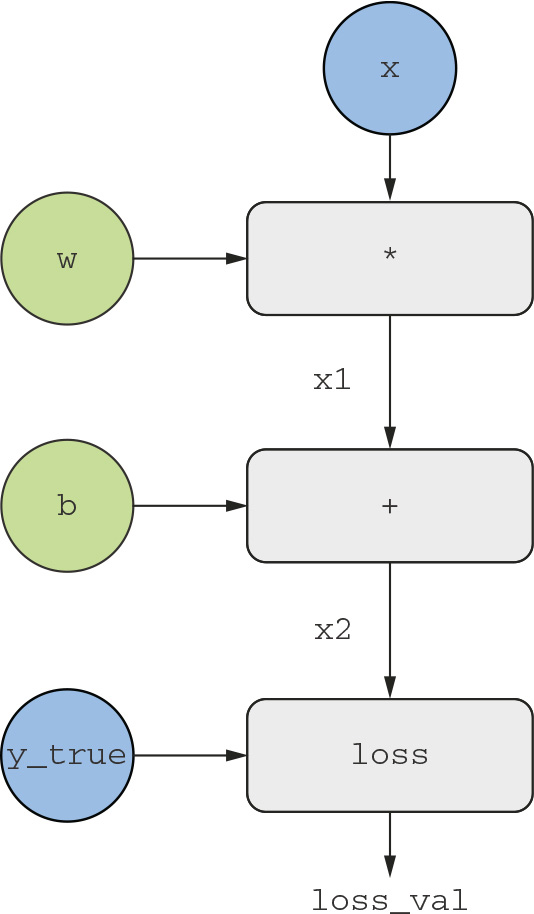
Figure 2.22 A basic example of a computation graph
Let’s set concrete values for the “input nodes” in the graph, that is to say, the input x, the target y_true, w, and b. We’ll propagate these values to all nodes in the graph, from top to bottom, until we reach loss_val. This is the forward pass (see figure 2.23).
Now let’s “reverse” the graph: for each edge in the graph going from A to B, we will create an opposite edge from B to A, and ask, how much does B vary when A varies? That is to say, what is grad(B, A)? We’ll annotate each inverted edge with this value. This backward graph represents the backward pass (see figure 2.24).
We have the following:
- grad(loss_val, x2) = 1, because as x2 varies by an amount epsilon, loss_val = abs (4 -x2) varies by the same amount.
- grad(x2, x1) = 1, because as x1 varies by an amount epsilon, x2 = x1 + b = x1 + 1 varies by the same amount.
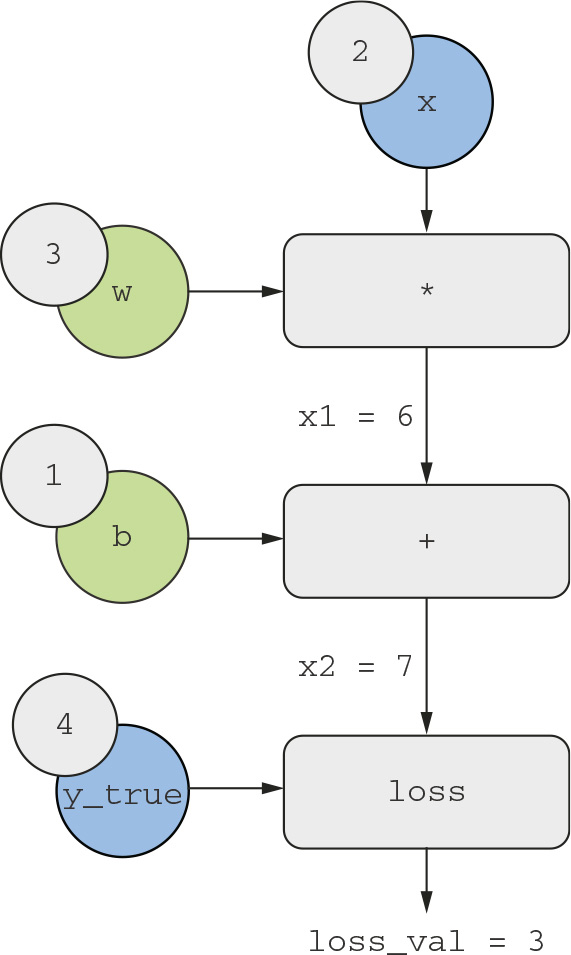
Figure 2.23 Running a forward pass

Figure 2.24 Running a backward pass
- grad(x2, b) = 1, because as b varies by an amount epsilon, x2 = x1 + b = 6 + b varies by the same amount.
- grad(x1, w) = 2, because as w varies by an amount epsilon, x1 = x * w = 2 * w varies by 2 * epsilon.
What the chain rule says about this backward graph is that you can obtain the derivative of a node with respect to another node by multiplying the derivatives for each edge along the path linking the two nodes, for instance, grad(loss_val, w) = grad(loss_val, x2) * grad(x2, x1) * grad(x1, w) (see figure 2.25).
By applying the chain rule to our graph, we obtain what we we’re looking for:
- grad(loss_val, w) = 1 * 1 * 2 = 2
- grad(loss_val, b) = 1 * 1 = 1
If there are multiple paths linking the two nodes of interest, a and b, in the backward graph, we would obtain grad(b, a) by summing the contributions of all the paths.
And with that, you just saw backpropagation in action! Backpropagation is simply the application of the chain rule to a computation graph. There’s nothing more to it. Backpropagation starts with the final loss value and works backward from the top layers to the bottom layers, computing the contribution that each parameter had in the loss value. That’s where the name “backpropagation” comes from: we “back-propagate” the loss contributions of different nodes in a computation graph.
Nowadays people implement neural networks in modern frameworks that are capable of automatic differentiation, such as TensorFlow. Automatic differentiation is implemented with the kind of computation graph you’ve just seen. Automatic differentiation makes it possible to retrieve the gradients of arbitrary compositions of differentiable tensor operations without doing any extra work besides writing down the forward pass. When I (François) wrote my first neural networks in C in the 2000s, I had to write my gradients by hand. Now, thanks to modern automatic differentiation tools, you’ll never have to implement backpropagation yourself. Consider yourself lucky!
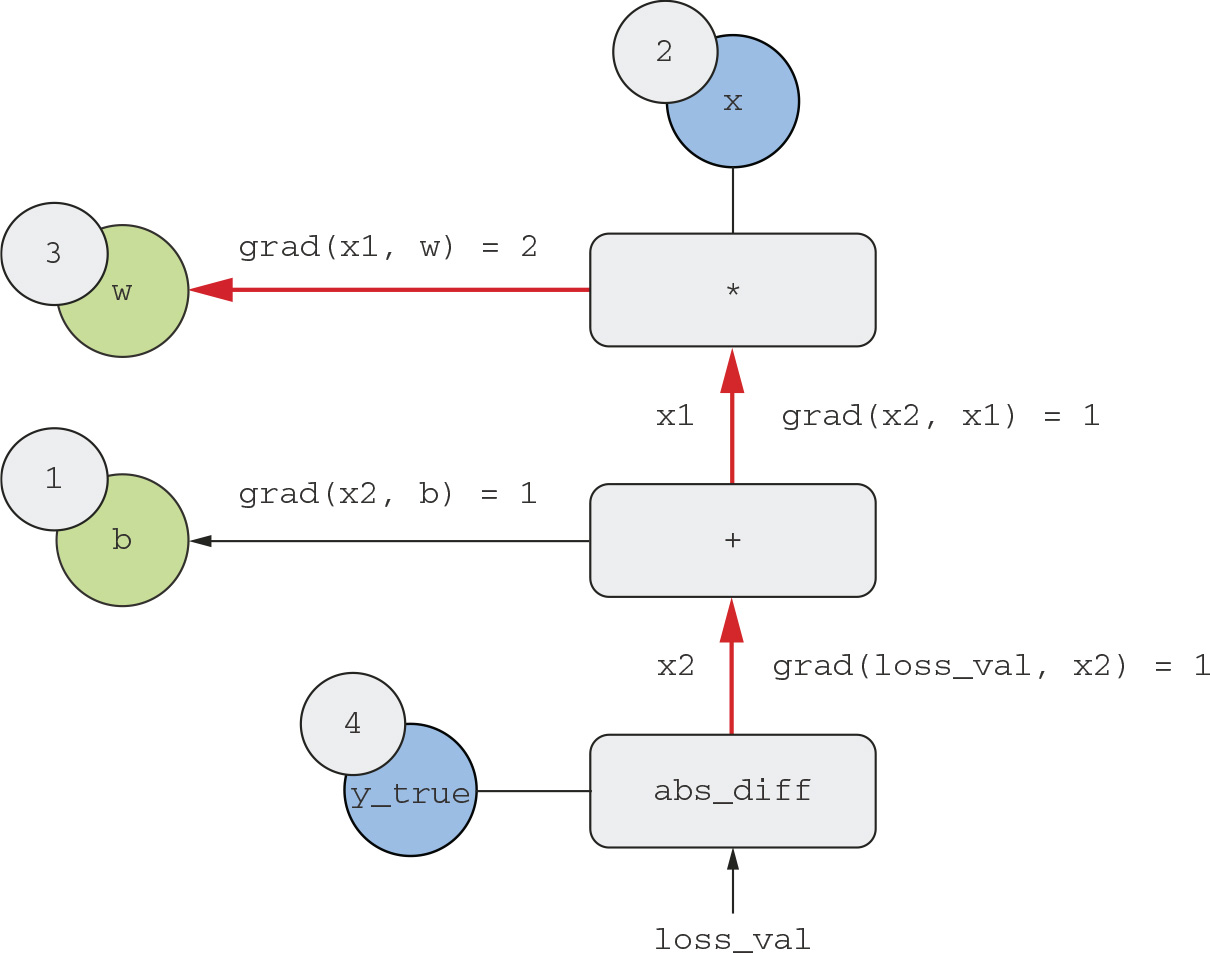
Figure 2.25 Path from loss_val to w in the backward graph
THE GRADIENT TAPE IN TENSORFLOW
The API through which you can leverage TensorFlow’s powerful automatic differentiation capabilities is the GradientTape(). It’s a context manager that will “record” the tensor operations that run inside its scope, in the form of a computation graph (sometimes called a “tape”). This graph can then be used to retrieve the gradient of any output with respect to any variable or set of variables (instances of the TensorFlow Variable class). A tf$Variable is a specific kind of tensor meant to hold mutable state—for instance, the weights of a neural network are always TensorFlow Variable instances:
library(tensorflow)
x <- tf$Variable(0)➊
with(tf$GradientTape() %as% tape, {➋
y <- 2 * x + 3➌
})➍
grad_of_y_wrt_x <- tape$gradient(y, x)➎
➊ Instantiate a scalar Variable with an initial value of 0.
➋ Open a GradientTape scope.
➌ Inside the scope, apply some tensor operations to our variable.
➍ Exit the scope.
➎ Use the tape to retrieve the gradient of the output y with respect to our variable x.
The GradientTape() works with tensor operations as follows:
x <- tf$Variable(array(0, dim = c(2, 2)))➊
with(tf$GradientTape() %as% tape, {
y <- 2 * x + 3
})
grad_of_y_wrt_x <- as.array(tape$gradient(y, x))➋
➊ Instantiate a variable with shape (2, 2) and an initial value of all zeros.
➋ grad_of_y_wrt_x is a tensor of shape (2, 2) (like x) describing the curvature of y = 2 * a + 3 around x = array(0, dim = c(2, 2)).
Note that tape$gradient() returns a TensorFlow Tensor, which we convert to an R array with as.array(). GradientTape() also works with lists of variables:
W <- tf$Variable(random_array(c(2, 2)))
b <- tf$Variable(array(0, dim = c(2)))
x <- random_array(c(2, 2))
with(tf$GradientTape() %as% tape, {
y <- tf$matmul(x, W) + b➊
})
grad_of_y_wrt_W_and_b <- tape$gradient(y, list(W, b))
str(grad_of_y_wrt_W_and_b)➋
List of 2
$ :<tf.Tensor: shape=(2, 2), dtype=float64, numpy=…>
$ :<tf.Tensor: shape=(2), dtype=float64, numpy=array([2., 2.])>
➊ matmul is how you say "dot product" in TensorFlow.
➋ grad_of_y_wrt_W_and_b is a list of two tensors with the same shapes as W and b, respectively.
You will learn more about the gradient tape in the next chapter.
2.5 Looking back at our first example
You’re nearing the end of this chapter, and you should now have a general understanding of what’s going on behind the scenes in a neural network. What was a magical black box at the start of the chapter has turned into a clearer picture, as illustrated in figure 2.26: the model, composed of layers that are chained together, maps the input data to predictions. The loss function then compares these predictions to the targets, producing a loss value: a measure of how well the model’s predictions match what was expected. The optimizer uses this loss value to update the model’s weights.

Figure 2.26 Relationship between the network, layers, loss function, and optimizer
Let’s go back to the first example in this chapter and review each piece of it in the light of what you’ve learned since.
This was the input data:
library(keras)
mnist <- dataset_mnist()
train_images <- mnist$train$x
train_images <- array_reshape(train_images, c(60000, 28 * 28))
train_images <- train_images / 255
test_images <- mnist$test$x
test_images <- array_reshape(test_images, c(10000, 28 * 28))
test_images <- test_images / 255
train_labels <- mnist$train$y
test_labels <- mnist$test$y
Now you understand that the input images are stored in R arrays of shape (60000, 784) (training data) and (10000, 784) (test data) respectively.
This was our model:
model <- keras_model_sequential(list(
layer_dense(units = 512, activation = "relu"),
layer_dense(units = 10, activation = "softmax")
))
Now you understand that this model consists of a chain of two Dense layers, that each layer applies a few simple tensor operations to the input data, and that these operations involve weight tensors. Weight tensors, which are attributes of the layers, are where the knowledge of the model persists.
This was the model-compilation step:
compile(model,
optimizer = "rmsprop",
loss = "sparse_categorical_crossentropy",
metrics = c("accuracy"))
Now you understand that sparse_categorical_crossentropy is the loss function that’s used as a feedback signal for learning the weight tensors and which the training phase will attempt to minimize. You also know that this reduction of the loss happens via mini-batch stochastic gradient descent. The exact rules governing a specific use of gradient descent are defined by the rmsprop optimizer passed as the first argument.
Finally, this was the training loop:
fit(model, train_images, train_labels, epochs = 5, batch_size = 128)
Now you understand what happens when you call fit: the model will start to iterate on the training data in mini-batches of 128 samples, five times over (each iteration over all the training data is called an epoch). For each batch, the model will compute the gradient of the loss with regard to the weights (using the backpropagation algorithm, which derives from the chain rule in calculus) and move the weights in the direction that will reduce the value of the loss for this batch.
After these five epochs, the model will have performed 2,345 gradient updates (469 per epoch), and the loss of the model will be sufficiently low that the model will be capable of classifying handwritten digits with high accuracy.
At this point, you already know most of what there is to know about neural networks. Let’s prove it by reimplementing a simplified version of that first example “from scratch” in TensorFlow, step by step.
2.5.1 Reimplementing our first example from scratch in TensorFlow
What better demonstrates full, unambiguous understanding than implementing everything from scratch? Of course, what “from scratch” means here is relative: we won’t reimplement basic tensor operations, and we won’t implement backpropagation. But we’ll go to such a low level that we will barely use any Keras functionality at all.
Don’t worry if you don’t understand every little detail in this example just yet. The next chapter will dive in more detail into the TensorFlow API. For now, just try to follow the gist of what’s going on—the intent of this example is to help crystallize your understanding of the mathematics of deep learning using a concrete implementation. Let’s go!
A SIMPLE DENSE CLASS
You’ve learned earlier that the Dense layer implements the following input transformation, where W and b are model parameters, and activation() is an element-wise function (usually relu(), but it would be softmax() for the last layer):
output <- activation(dot(W, input) + b)
Let’s implement a simple Dense layer as a plain R environment with a class attribute NaiveDense, two TensorFlow variables, W and b, and a call() method that applies the preceding transformation:
layer_naive_dense <- function(input_size, output_size, activation) {
self <- new.env(parent = emptyenv())
attr(self, "class") <- "NaiveDense"
self$activation <- activation
w_shape <- c(input_size, output_size)
w_initial_value <- random_array(w_shape, min = 0, max = 1e-1)
self$W <- tf$Variable(w_initial_value)➊
b_shape <- c(output_size)
b_initial_value <- array(0, b_shape)
self$b <- tf$Variable(b_initial_value)➋
self$weights <- list(self$W, self$b)➌
self$call <- function(inputs) {➍
self$activation(tf$matmul(inputs, self$W) + self$b)➎
}
self
}
➊ Create a matrix, W, of shape (input_size, output_size), initialized with random values.
➋ Create a vector, b, of shape (output_size), initialized with zeros.
➌ Convenience property for retrieving all the layer's weights
➍ Apply the forward pass in a function named call.
➎ We stick to TensorFlow operations in this function, so that GradientTape can track them. (We'll learn more about TensorFlow operations in chapter 3.)
A SIMPLE SEQUENTIAL CLASS
Now, let’s create a naive_model_sequential() to chain these layers, as shown in the next code snippet. It wraps a list of layers and exposes a call() method that simply calls the underlying layers on the inputs, in order. It also features a weights property to easily keep track of the layers’ parameters:
naive_model_sequential <- function(layers) {
self <- new.env(parent = emptyenv())
attr(self, "class") <- "NaiveSequential"
self$layers <- layers
weights <- lapply(layers, function(layer) layer$weights)
self$weights <- do.call(c, weights)➊
self$call <- function(inputs) {
x <- inputs
for (layer in self$layers)
x <- layer$call(x)
x
}
self
}
➊ Flatten the nested list one level.
Using this NaiveDense class and this NaiveSequential class, we can create a mock Keras model:
model <- naive_model_sequential(list(
layer_naive_dense(input_size = 28 * 28, output_size = 512,
activation = tf$nn$relu),
layer_naive_dense(input_size = 512, output_size = 10,
activation = tf$nn$softmax)
))
stopifnot(length(model$weights) == 4)
A BATCH GENERATOR
Next, we need a way to iterate over the MNIST data in mini-batches. This is easy:
new_batch_generator <- function(images, labels, batch_size = 128) {
self <- new.env(parent = emptyenv())
attr(self, "class") <- "BatchGenerator"
stopifnot(nrow(images) == nrow(labels))
self$index <- 1
self$images <- images
self$labels <- labels self$batch_size <- batch_size
self$num_batches <- ceiling(nrow(images) / batch_size)
self$get_next_batch <- function() {
start <- self$index
if(start > nrow(images))
return(NULL)➊
end <- start + self$batch_size - 1
if(end > nrow(images))
end <- nrow(images)➋
self$index <- end + 1
indices <- start:end
list(images = self$images[indices, ],
labels = self$labels[indices])
}
self
}
➊ Generator is finished.
➋ Last batch may be smaller.
2.5.2 Running one training step
The most difficult part of the process is the “training step”: updating the weights of the model after running it on one batch of data. We need to do the following:
- 1 Compute the predictions of the model for the images in the batch.
- 2 Compute the loss value for these predictions, given the actual labels.
- 3 Compute the gradient of the loss with regard to the model’s weights.
- 4 Move the weights by a small amount in the direction opposite to the gradient.
To compute the gradient, we will use the TensorFlow GradientTape object we introduced in section 2.4.4:
one_training_step <- function(model, images_batch, labels_batch) {
with(tf$GradientTape() %as% tape, {
predictions <- model$call(images_batch)➊
per_sample_losses <
loss_sparse_categorical_crossentropy(labels_batch, predictions)
average_loss <- mean(per_sample_losses)
})
gradients <- tape$gradient(average_loss, model$weights)➋
update_weights(gradients, model$weights)➌
average_loss
}
➊ Run the forward pass (compute the model's predictions under a GradientTape scope).
➋ Compute the gradient of the loss with regard to the weights. The output gradients is a list where each entry corresponds to a weight from the model$weights list.
➌ Update the weights using the gradients (we will define this function shortly).
As you already know, the purpose of the “weight update” step (represented by the preceding update_weights() function) is to move the weights by “a bit” in a direction that will reduce the loss on this batch. The magnitude of the move is determined by the “learning rate,” typically a small quantity. The simplest way to implement this update_ weights() function is to subtract gradient * learning_rate from each weight:
learning_rate <- 1e-3
update_weights <- function(gradients, weights) {
stopifnot(length(gradients) == length(weights))
for (i in seq_along(weights))
weights[[i]]$assign_sub(➊
gradients[[i]] * learning_rate)
}
➊ x$assign_sub(value) is the equivalent of x <- x - value for TensorFlow variables.
In practice, you would almost never implement a weight update step like this by hand. Instead, you would use an Optimizer instance from Keras:
optimizer <- optimizer_sgd(learning_rate = 1e-3)
update_weights <- function(gradients, weights)
optimizer$apply_gradients(zip_lists(gradients, weights))
zip_lists() is a helper function that we use to turn the lists of gradients and weights into a list of (gradient, weight) pairs. We use it to pair gradients with weights for the optimizer. For example:
str(zip_lists(
gradients = list("grad_for_wt_1", "grad_for_wt_2", "grad_for_wt_3"),
weights = list("weight_1", "weight_2", "weight_3")))
List of 3
$ :List of 2
..$ gradients: chr "grad_for_wt_1"
..$ weights : chr "weight_1"
$ :List of 2
..$ gradients: chr "grad_for_wt_2"
..$ weights : chr "weight_2"
$ :List of 2
..$ gradients: chr "grad_for_wt_3"
..$ weights : chr "weight_3"
Now that our per-batch training step is ready, we can move on to implementing an entire epoch of training.
2.5.3 The full training loop
An epoch of training simply consists of repeating the training step for each batch in the training data, and the full training loop is simply the repetition of one epoch:
fit <- function(model, images, labels, epochs, batch_size = 128) {
for (epoch_counter in seq_len(epochs)) {
cat("Epoch ", epoch_counter, " ")
batch_generator <- new_batch_generator(images, labels)
for (batch_counter in seq_len(batch_generator$num_batches)) {
batch <- batch_generator$get_next_batch()
loss <- one_training_step(model, batch$images, batch$labels)
if (batch_counter %% 100 == 0)
cat(sprintf("loss at batch %s: %.2f ", batch_counter, loss))
}
}
}
Let’s test-drive it:
mnist <- dataset_mnist()
train_images <- array_reshape(mnist$train$x, c(60000, 28 * 28)) / 255
test_images <- array_reshape(mnist$test$x, c(10000, 28 * 28)) / 255
test_labels <- mnist$test$y
train_labels <- mnist$train$y
fit(model, train_images, train_labels, epochs = 10, batch_size = 128)
Epoch 1
loss at batch 100: 2.37
loss at batch 200: 2.21
loss at batch 300: 2.15
loss at batch 400: 2.09
Epoch 2
loss at batch 100: 1.98
loss at batch 200: 1.83
loss at batch 300: 1.83
loss at batch 400: 1.75
…
Epoch 9
loss at batch 100: 0.85
loss at batch 200: 0.68
loss at batch 300: 0.83
loss at batch 400: 0.76
Epoch 10
loss at batch 100: 0.80
loss at batch 200: 0.63
loss at batch 300: 0.78
loss at batch 400: 0.72
2.5.4 Evaluating the model
We can evaluate the model by taking the max.col() of its predictions over the test images, and comparing it to the expected labels:
predictions <- model$call(test_images)
predictions <- as.array(predictions)➊
predicted_labels <- max.col(predictions) - 1➋ ➌
matches <- predicted_labels == test_labels
cat(sprintf("accuracy: %.2f ", mean(matches)))
accuracy: 0.82
➊ Convert the TensorFlow Tensor to an R array.
➋ max.col(x) is a vectorized implementation of apply(x, 1, which.max)).
➌ Subtract 1 because positions are offset from labels by 1, for example, the first position corresponds to digit 0.
All done! As you can see, it’s quite a bit of work to do “by hand” what you can do in a few lines of Keras code. But because you’ve gone through these steps, you should now have a crystal-clear understanding of what goes on inside a neural network when you call fit(). Having this low-level mental model of what your code is doing behind the scenes will make you better able to leverage the high-level features of the Keras API.
Summary
- Tensors form the foundation of modern machine learning systems. They come in various flavors of type, rank, and shape.
- You can manipulate numerical tensors via tensor operations (such as addition, tensor product, or element-wise multiplication), which can be interpreted as encoding geometric transformations. In general, everything in deep learning is amenable to a geometric interpretation.
- Deep learning models consist of chains of simple tensor operations, parameterized by weights, which are themselves tensors. The weights of a model are where its “knowledge” is stored.
- Learning means finding a set of values for the model’s weights that minimizes a loss function for a given set of training data samples and their corresponding targets.
- Learning happens by drawing random batches of data samples and their targets and computing the gradient of the model parameters with respect to the loss on the batch. The model parameters are then moved a bit (the magnitude of the move is defined by the learning rate) in the opposite direction from the gradient. This is called mini-batch stochastic gradient descent.
- The entire learning process is made possible by the fact that all tensor operations in neural networks are differentiable, and thus it’s possible to apply the chain rule of derivation to find the gradient function mapping the current parameters and current batch of data to a gradient value. This is called back-propagation.
- Two key concepts you’ll see frequently in future chapters are loss and optimizer. These are the two things you need to define before you begin feeding data into a model:
- The loss is the quantity you’ll attempt to minimize during training, so it should represent a measure of success for the task you’re trying to solve.
- The optimizer specifies the exact way in which the gradient of the loss will be used to update parameters: for instance, it could be the RMSprop optimizer, SGD with momentum, and so on.
