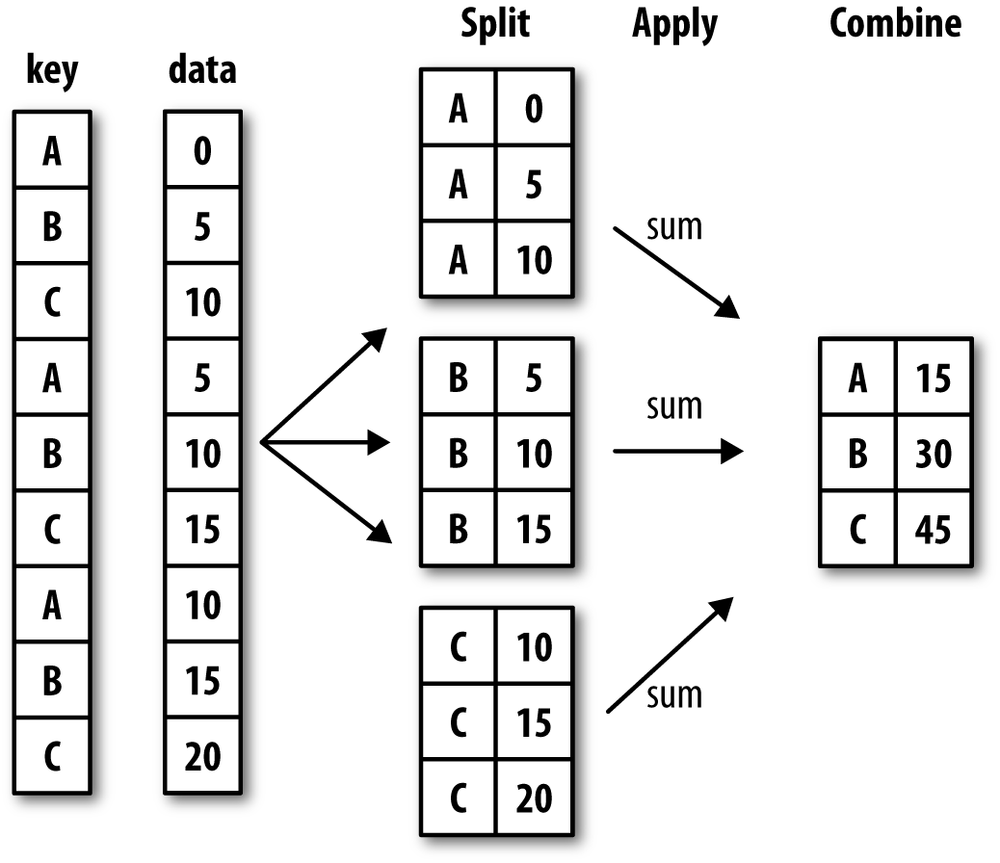
Categorizing a data set and applying a function to each group, whether
an aggregation or transformation, is often a critical component of a data
analysis workflow. After loading, merging, and preparing a data set, a
familiar task is to compute group statistics or possibly pivot
tables for reporting or visualization purposes. pandas provides a
flexible and high-performance groupby
facility, enabling you to slice and dice, and summarize data sets in a
natural way.
One reason for the popularity of relational databases and SQL (which stands for “structured query language”) is the ease with which data can be joined, filtered, transformed, and aggregated. However, query languages like SQL are rather limited in the kinds of group operations that can be performed. As you will see, with the expressiveness and power of Python and pandas, we can perform much more complex grouped operations by utilizing any function that accepts a pandas object or NumPy array. In this chapter, you will learn how to:
Split a pandas object into pieces using one or more keys (in the form of functions, arrays, or DataFrame column names)
Computing group summary statistics, like count, mean, or standard deviation, or a user-defined function
Apply a varying set of functions to each column of a DataFrame
Apply within-group transformations or other manipulations, like normalization, linear regression, rank, or subset selection
Compute pivot tables and cross-tabulations
Perform quantile analysis and other data-derived group analyses
Note
Aggregation of time series data, a special use case of groupby, is referred to as
resampling in this book and will receive separate
treatment in Chapter 10.
Hadley Wickham, an author of many popular packages for the R
programming language, coined the term split-apply-combine for talking about
group operations, and I think that’s a good description of the process. In
the first stage of the process, data contained in a pandas object, whether
a Series, DataFrame, or otherwise, is split into
groups based on one or more keys that you provide.
The splitting is performed on a particular axis of an object. For example,
a DataFrame can be grouped on its rows (axis=0) or its columns (axis=1). Once this is done, a function is
applied to each group, producing a new value.
Finally, the results of all those function applications are
combined into a result object. The form of the
resulting object will usually depend on what’s being done to the data. See
Figure 9-1 for a mockup of a simple group
aggregation.
Each grouping key can take many forms, and the keys do not have to be all of the same type:
A list or array of values that is the same length as the axis being grouped
A value indicating a column name in a DataFrame
A dict or Series giving a correspondence between the values on the axis being grouped and the group names
A function to be invoked on the axis index or the individual labels in the index
Note that the latter three methods are all just shortcuts for producing an array of values to be used to split up the object. Don’t worry if this all seems very abstract. Throughout this chapter, I will give many examples of all of these methods. To get started, here is a very simple small tabular dataset as a DataFrame:
In [160]: df = DataFrame({'key1' : ['a', 'a', 'b', 'b', 'a'],
.....: 'key2' : ['one', 'two', 'one', 'two', 'one'],
.....: 'data1' : np.random.randn(5),
.....: 'data2' : np.random.randn(5)})
In [161]: df
Out[161]:
data1 data2 key1 key2
0 -0.204708 1.393406 a one
1 0.478943 0.092908 a two
2 -0.519439 0.281746 b one
3 -0.555730 0.769023 b two
4 1.965781 1.246435 a oneSuppose you wanted to compute the mean of the data1 column using the groups labels from
key1. There are a number of ways to do
this. One is to access data1 and call
groupby with the column (a Series) at
key1:
In [162]: grouped = df['data1'].groupby(df['key1']) In [163]: grouped Out[163]: <pandas.core.groupby.SeriesGroupBy object at 0x7f5893ec7750>
This grouped variable is now a
GroupBy object. It has not actually computed anything
yet except for some intermediate data about the group key df['key1']. The idea is that this object has all
of the information needed to then apply some operation to each of the
groups. For example, to compute group means we can call the GroupBy’s
mean method:
In [164]: grouped.mean() Out[164]: key1 a 0.746672 b -0.537585 Name: data1, dtype: float64
Later, I’ll explain more about what’s going on when you call
.mean(). The important
thing here is that the data (a Series) has been aggregated according to
the group key, producing a new Series that is now indexed by the unique
values in the key1 column. The result
index has the name 'key1' because the
DataFrame column df['key1'] did.
If instead we had passed multiple arrays as a list, we get something different:
In [165]: means = df['data1'].groupby([df['key1'], df['key2']]).mean()
In [166]: means
Out[166]:
key1 key2
a one 0.880536
two 0.478943
b one -0.519439
two -0.555730
Name: data1, dtype: float64In this case, we grouped the data using two keys, and the resulting Series now has a hierarchical index consisting of the unique pairs of keys observed:
In [167]: means.unstack() Out[167]: key2 one two key1 a 0.880536 0.478943 b -0.519439 -0.555730
In these examples, the group keys are all Series, though they could be any arrays of the right length:
In [168]: states = np.array(['Ohio', 'California', 'California', 'Ohio', 'Ohio'])
In [169]: years = np.array([2005, 2005, 2006, 2005, 2006])
In [170]: df['data1'].groupby([states, years]).mean()
Out[170]:
California 2005 0.478943
2006 -0.519439
Ohio 2005 -0.380219
2006 1.965781
Name: data1, dtype: float64Frequently the grouping information to be found in the same DataFrame as the data you want to work on. In that case, you can pass column names (whether those are strings, numbers, or other Python objects) as the group keys:
In [171]: df.groupby('key1').mean()
Out[171]:
data1 data2
key1
a 0.746672 0.910916
b -0.537585 0.525384
In [172]: df.groupby(['key1', 'key2']).mean()
Out[172]:
data1 data2
key1 key2
a one 0.880536 1.319920
two 0.478943 0.092908
b one -0.519439 0.281746
two -0.555730 0.769023You may have noticed in the first case df.groupby('key1').mean() that there is no
key2 column in the result. Because
df['key2'] is not numeric data, it is
said to be a nuisance column, which is therefore
excluded from the result. By default, all of the numeric columns are aggregated, though it is
possible to filter down to a subset as you’ll see soon.
Regardless of the objective in using groupby, a generally useful GroupBy method is
size which return a
Series containing group sizes:
In [173]: df.groupby(['key1', 'key2']).size()
Out[173]:
key1 key2
a one 2
two 1
b one 1
two 1
dtype: int64Caution
As of this writing, any missing values in a group key will be
excluded from the result. It’s possible (and, in fact, quite likely),
that by the time you are reading this there will be an option to include
the NA group in the result.
The GroupBy object supports iteration, generating a sequence of 2-tuples containing the group name along with the chunk of data. Consider the following small example data set:
In [174]: for name, group in df.groupby('key1'):
.....: print(name)
.....: print(group)
.....:
a
data1 data2 key1 key2
0 -0.204708 1.393406 a one
1 0.478943 0.092908 a two
4 1.965781 1.246435 a one
b
data1 data2 key1 key2
2 -0.519439 0.281746 b one
3 -0.555730 0.769023 b twoIn the case of multiple keys, the first element in the tuple will be a tuple of key values:
In [175]: for (k1, k2), group in df.groupby(['key1', 'key2']):
.....: print((k1, k2))
.....: print(group)
.....:
a one
data1 data2 key1 key2
0 -0.204708 1.393406 a one
4 1.965781 1.246435 a one
a two
data1 data2 key1 key2
1 0.478943 0.092908 a two
b one
data1 data2 key1 key2
2 -0.519439 0.281746 b one
b two
data1 data2 key1 key2
3 -0.55573 0.769023 b twoOf course, you can choose to do whatever you want with the pieces of data. A recipe you may find useful is computing a dict of the data pieces as a one-liner:
In [176]: pieces = dict(list(df.groupby('key1')))
In [177]: pieces['b']
Out[177]:
data1 data2 key1 key2
2 -0.519439 0.281746 b one
3 -0.555730 0.769023 b twoBy default groupby groups on
axis=0, but you can group on any of
the other axes. For example, we could group the columns of our example
df here by dtype like so:
In [178]: df.dtypes
Out[178]:
data1 float64
data2 float64
key1 object
key2 object
dtype: object
In [179]: grouped = df.groupby(df.dtypes, axis=1)
In [180]: dict(list(grouped))
Out[180]:
{dtype('float64'): data1 data2
0 -0.204708 1.393406
1 0.478943 0.092908
2 -0.519439 0.281746
3 -0.555730 0.769023
4 1.965781 1.246435, dtype('O'): key1 key2
0 a one
1 a two
2 b one
3 b two
4 a one}Indexing a GroupBy object created from a DataFrame with a column name or array of column names has the effect of selecting those columns for aggregation. This means that:
df.groupby('key1')['data1']
df.groupby('key1')[['data2']]are syntactic sugar for:
df['data1'].groupby(df['key1']) df[['data2']].groupby(df['key1'])
Especially for large data sets, it may be desirable to aggregate
only a few columns. For example, in the above data set, to compute means
for just the data2 column and get the
result as a DataFrame, we could write:
In [181]: df.groupby(['key1', 'key2'])[['data2']].mean()
Out[181]:
data2
key1 key2
a one 1.319920
two 0.092908
b one 0.281746
two 0.769023The object returned by this indexing operation is a grouped DataFrame if a list or array is passed and a grouped Series is just a single column name that is passed as a scalar:
In [182]: s_grouped = df.groupby(['key1', 'key2'])['data2']
In [183]: s_grouped
Out[183]: <pandas.core.groupby.SeriesGroupBy object at 0x7f5893e77890>
In [184]: s_grouped.mean()
Out[184]:
key1 key2
a one 1.319920
two 0.092908
b one 0.281746
two 0.769023
Name: data2, dtype: float64Grouping information may exist in a form other than an array. Let’s consider another example DataFrame:
In [185]: people = DataFrame(np.random.randn(5, 5),
.....: columns=['a', 'b', 'c', 'd', 'e'],
.....: index=['Joe', 'Steve', 'Wes', 'Jim', 'Travis'])
In [186]: people.ix[2:3, ['b', 'c']] = np.nan # Add a few NA values
In [187]: people
Out[187]:
a b c d e
Joe 1.007189 -1.296221 0.274992 0.228913 1.352917
Steve 0.886429 -2.001637 -0.371843 1.669025 -0.438570
Wes -0.539741 NaN NaN -1.021228 -0.577087
Jim 0.124121 0.302614 0.523772 0.000940 1.343810
Travis -0.713544 -0.831154 -2.370232 -1.860761 -0.860757Now, suppose I have a group correspondence for the columns and want to sum together the columns by group:
In [188]: mapping = {'a': 'red', 'b': 'red', 'c': 'blue',
.....: 'd': 'blue', 'e': 'red', 'f' : 'orange'}Now, you could easily construct an array from this dict to pass to
groupby, but instead we can just pass
the dict:
In [189]: by_column = people.groupby(mapping, axis=1)
In [190]: by_column.sum()
Out[190]:
blue red
Joe 0.503905 1.063885
Steve 1.297183 -1.553778
Wes -1.021228 -1.116829
Jim 0.524712 1.770545
Travis -4.230992 -2.405455The same functionality holds for Series, which can be viewed as a fixed size mapping. When I used Series as group keys in the above examples, pandas does, in fact, inspect each Series to ensure that its index is aligned with the axis it’s grouping:
In [191]: map_series = Series(mapping)
In [192]: map_series
Out[192]:
a red
b red
c blue
d blue
e red
f orange
dtype: object
In [193]: people.groupby(map_series, axis=1).count()
Out[193]:
blue red
Joe 2 3
Steve 2 3
Wes 1 2
Jim 2 3
Travis 2 3Using Python functions in what can be fairly creative ways
is a more abstract way of defining a group mapping compared with a dict
or Series. Any function passed as a group key will be called once per
index value, with the return values being used as the group names. More
concretely, consider the example DataFrame from the previous section,
which has people’s first names as index values. Suppose you wanted to
group by the length of the names; you could compute an array of string
lengths, but instead you can just pass the len function:
In [194]: people.groupby(len).sum()
Out[194]:
a b c d e
3 0.591569 -0.993608 0.798764 -0.791374 2.119639
5 0.886429 -2.001637 -0.371843 1.669025 -0.438570
6 -0.713544 -0.831154 -2.370232 -1.860761 -0.860757Mixing functions with arrays, dicts, or Series is not a problem as everything gets converted to arrays internally:
In [195]: key_list = ['one', 'one', 'one', 'two', 'two']
In [196]: people.groupby([len, key_list]).min()
Out[196]:
a b c d e
3 one -0.539741 -1.296221 0.274992 -1.021228 -0.577087
two 0.124121 0.302614 0.523772 0.000940 1.343810
5 one 0.886429 -2.001637 -0.371843 1.669025 -0.438570
6 two -0.713544 -0.831154 -2.370232 -1.860761 -0.860757A final convenience for hierarchically-indexed data sets
is the ability to aggregate using one of the levels of an axis index. To
do this, pass the level number or name using the level
keyword:
In [197]: columns = pd.MultiIndex.from_arrays([['US', 'US', 'US', 'JP', 'JP'], .....: [1, 3, 5, 1, 3]], names=['cty', 'tenor']) In [198]: hier_df = DataFrame(np.random.randn(4, 5), columns=columns) In [199]: hier_df Out[199]: cty US JP tenor 1 3 5 1 3 0 0.560145 -1.265934 0.119827 -1.063512 0.332883 1 -2.359419 -0.199543 -1.541996 -0.970736 -1.307030 2 0.286350 0.377984 -0.753887 0.331286 1.349742 3 0.069877 0.246674 -0.011862 1.004812 1.327195 In [200]: hier_df.groupby(level='cty', axis=1).count() Out[200]: cty JP US 0 2 3 1 2 3 2 2 3 3 2 3
By aggregation, I am generally referring to any data
transformation that produces scalar values from arrays. In the examples
above I have used several of them, such as mean,
count, min and sum. You may wonder what
is going on when you invoke mean() on a GroupBy
object. Many common aggregations, such as those found in Table 9-1, have optimized implementations
that compute the statistics on the dataset in place.
However, you are not limited to only this set of methods. You can use
aggregations of your own devising and additionally call any method that is
also defined on the grouped object. For example, as you recall quantile computes sample quantiles of a Series
or a DataFrame’s columns [3]:
In [201]: df
Out[201]:
data1 data2 key1 key2
0 -0.204708 1.393406 a one
1 0.478943 0.092908 a two
2 -0.519439 0.281746 b one
3 -0.555730 0.769023 b two
4 1.965781 1.246435 a one
In [202]: grouped = df.groupby('key1')
In [203]: grouped['data1'].quantile(0.9)
Out[203]:
key1
a 1.668413
b -0.523068
Name: data1, dtype: float64While quantile is not explicitly
implemented for GroupBy, it is a Series method and thus available for use.
Internally, GroupBy efficiently slices up the Series, calls piece.quantile(0.9) for each piece, then
assembles those results together into the result object.
To use your own aggregation functions, pass any function that
aggregates an array to the aggregate or agg method:
In [204]: def peak_to_peak(arr):
.....: return arr.max() - arr.min()
In [205]: grouped.agg(peak_to_peak)
Out[205]:
data1 data2
key1
a 2.170488 1.300498
b 0.036292 0.487276You’ll notice that some methods like describe also work, even though they are not
aggregations, strictly speaking:
In [206]: grouped.describe()
Out[206]:
data1 data2
key1
a count 3.000000 3.000000
mean 0.746672 0.910916
std 1.109736 0.712217
min -0.204708 0.092908
25% 0.137118 0.669671
... ... ...
b min -0.555730 0.281746
25% -0.546657 0.403565
50% -0.537585 0.525384
75% -0.528512 0.647203
max -0.519439 0.769023
[16 rows x 2 columns]I will explain in more detail what has happened here in the next major section on group-wise operations and transformations.
Note
You may notice that custom aggregation functions are much slower than the optimized functions found in Table 9-1. This is because there is significant overhead (function calls, data rearrangement) in constructing the intermediate group data chunks.
Table 9-1. Optimized groupby methods
To illustrate some more advanced aggregation features, I’ll use a
less trivial dataset, a dataset on restaurant tipping. I obtained it from
the R reshape2 package; it was
originally found in Bryant & Smith’s 1995 text on business statistics
(and found in the book’s GitHub repository). After loading it with
read_csv, I add a tipping
percentage column tip_pct.
In [207]: tips = pd.read_csv('ch08/tips.csv')
# Add tip percentage of total bill
In [208]: tips['tip_pct'] = tips['tip'] / tips['total_bill']
In [209]: tips[:6]
Out[209]:
total_bill tip sex smoker day time size tip_pct
0 16.99 1.01 Female No Sun Dinner 2 0.059447
1 10.34 1.66 Male No Sun Dinner 3 0.160542
2 21.01 3.50 Male No Sun Dinner 3 0.166587
3 23.68 3.31 Male No Sun Dinner 2 0.139780
4 24.59 3.61 Female No Sun Dinner 4 0.146808
5 25.29 4.71 Male No Sun Dinner 4 0.186240As you’ve seen above, aggregating a Series or all of the
columns of a DataFrame is a matter of using aggregate with the
desired function or calling a method like mean or std. However, you may want to aggregate using
a different function depending on the column or multiple functions at
once. Fortunately, this is straightforward to do, which I’ll illustrate
through a number of examples. First, I’ll group the tips by sex
and smoker:
In [210]: grouped = tips.groupby(['sex', 'smoker'])
Note that for descriptive statistics like those in Table 9-1, you can pass the name of the function as a string:
In [211]: grouped_pct = grouped['tip_pct']
In [212]: grouped_pct.agg('mean')
Out[212]:
sex smoker
Female No 0.156921
Yes 0.182150
Male No 0.160669
Yes 0.152771
Name: tip_pct, dtype: float64If you pass a list of functions or function names instead, you get back a DataFrame with column names taken from the functions:
In [213]: grouped_pct.agg(['mean', 'std', peak_to_peak])
Out[213]:
mean std peak_to_peak
sex smoker
Female No 0.156921 0.036421 0.195876
Yes 0.182150 0.071595 0.360233
Male No 0.160669 0.041849 0.220186
Yes 0.152771 0.090588 0.674707You don’t need to accept the names that GroupBy gives to the
columns; notably lambda functions have
the name '<lambda>' which make
them hard to identify (you can see for yourself by looking at a
function’s __name__ attribute). As
such, if you pass a list of (name,
function) tuples, the first element of each tuple will be used
as the DataFrame column names (you can think of a list of 2-tuples as an
ordered mapping):
In [214]: grouped_pct.agg([('foo', 'mean'), ('bar', np.std)])
Out[214]:
foo bar
sex smoker
Female No 0.156921 0.036421
Yes 0.182150 0.071595
Male No 0.160669 0.041849
Yes 0.152771 0.090588With a DataFrame, you have more options as you can specify a list
of functions to apply to all of the columns or different functions per
column. To start, suppose we wanted to compute the same three statistics
for the tip_pct and total_bill columns:
In [215]: functions = ['count', 'mean', 'max']
In [216]: result = grouped['tip_pct', 'total_bill'].agg(functions)
In [217]: result
Out[217]:
tip_pct total_bill
count mean max count mean max
sex smoker
Female No 54 0.156921 0.252672 54 18.105185 35.83
Yes 33 0.182150 0.416667 33 17.977879 44.30
Male No 97 0.160669 0.291990 97 19.791237 48.33
Yes 60 0.152771 0.710345 60 22.284500 50.81As you can see, the resulting DataFrame has hierarchical columns,
the same as you would get aggregating each column separately and using
concat to glue the results together
using the column names as the keys
argument:
In [218]: result['tip_pct']
Out[218]:
count mean max
sex smoker
Female No 54 0.156921 0.252672
Yes 33 0.182150 0.416667
Male No 97 0.160669 0.291990
Yes 60 0.152771 0.710345As above, a list of tuples with custom names can be passed:
In [219]: ftuples = [('Durchschnitt', 'mean'), ('Abweichung', np.var)]
In [220]: grouped['tip_pct', 'total_bill'].agg(ftuples)
Out[220]:
tip_pct total_bill
Durchschnitt Abweichung Durchschnitt Abweichung
sex smoker
Female No 0.156921 0.001327 18.105185 53.092422
Yes 0.182150 0.005126 17.977879 84.451517
Male No 0.160669 0.001751 19.791237 76.152961
Yes 0.152771 0.008206 22.284500 98.244673Now, suppose you wanted to apply potentially different functions
to one or more of the columns. The trick is to pass a dict to agg that contains a mapping of column names to
any of the function specifications listed so far:
In [221]: grouped.agg({'tip' : np.max, 'size' : 'sum'})
Out[221]:
tip size
sex smoker
Female No 5.2 140
Yes 6.5 74
Male No 9.0 263
Yes 10.0 150
In [222]: grouped.agg({'tip_pct' : ['min', 'max', 'mean', 'std'],
.....: 'size' : 'sum'})
Out[222]:
tip_pct size
min max mean std sum
sex smoker
Female No 0.056797 0.252672 0.156921 0.036421 140
Yes 0.056433 0.416667 0.182150 0.071595 74
Male No 0.071804 0.291990 0.160669 0.041849 263
Yes 0.035638 0.710345 0.152771 0.090588 150A DataFrame will have hierarchical columns only if multiple functions are applied to at least one column.
In all of the examples up until now, the aggregated data
comes back with an index, potentially hierarchical, composed from the
unique group key combinations observed. Since this isn’t always
desirable, you can disable this behavior in most cases by passing
as_index=False to groupby:
In [223]: tips.groupby(['sex', 'smoker'], as_index=False).mean()
Out[223]:
sex smoker total_bill tip size tip_pct
0 Female No 18.105185 2.773519 2.592593 0.156921
1 Female Yes 17.977879 2.931515 2.242424 0.182150
2 Male No 19.791237 3.113402 2.711340 0.160669
3 Male Yes 22.284500 3.051167 2.500000 0.152771Of course, it’s always possible to obtain the result in this
format by calling reset_index on the
result.
Aggregation is only one kind of group operation. It is a special
case in the more general class of data transformations; that is, it
accepts functions that reduce a one-dimensional array to a scalar value.
In this section, I will introduce you to the transform and apply methods, which will enable you to do many
other kinds of group operations.
Suppose, instead, we wanted to add a column to a DataFrame containing group means for each index. One way to do this is to aggregate, then merge:
In [224]: df
Out[224]:
data1 data2 key1 key2
0 -0.204708 1.393406 a one
1 0.478943 0.092908 a two
2 -0.519439 0.281746 b one
3 -0.555730 0.769023 b two
4 1.965781 1.246435 a one
In [225]: k1_means = df.groupby('key1').mean().add_prefix('mean_')
In [226]: k1_means
Out[226]:
mean_data1 mean_data2
key1
a 0.746672 0.910916
b -0.537585 0.525384
In [227]: pd.merge(df, k1_means, left_on='key1', right_index=True)
Out[227]:
data1 data2 key1 key2 mean_data1 mean_data2
0 -0.204708 1.393406 a one 0.746672 0.910916
1 0.478943 0.092908 a two 0.746672 0.910916
4 1.965781 1.246435 a one 0.746672 0.910916
2 -0.519439 0.281746 b one -0.537585 0.525384
3 -0.555730 0.769023 b two -0.537585 0.525384This works, but is somewhat inflexible. You can think of the
operation as transforming the two data columns using the np.mean function. Let’s
look back at the people DataFrame from
earlier in the chapter and use the transform method on GroupBy:
In [228]: key = ['one', 'two', 'one', 'two', 'one']
In [229]: people.groupby(key).mean()
Out[229]:
a b c d e
one -0.082032 -1.063687 -1.047620 -0.884358 -0.028309
two 0.505275 -0.849512 0.075965 0.834983 0.452620
In [230]: people.groupby(key).transform(np.mean)
Out[230]:
a b c d e
Jim NaN NaN NaN NaN NaN
Joe NaN NaN NaN NaN NaN
Steve NaN NaN NaN NaN NaN
Travis NaN NaN NaN NaN NaN
Wes NaN NaN NaN NaN NaN
one -0.082032 -1.063687 -1.047620 -0.884358 -0.028309
two 0.505275 -0.849512 0.075965 0.834983 0.452620As you may guess, transform
applies a function to each group, then places the results in the
appropriate locations. If each group produces a scalar value, it will be
propagated (broadcasted). Suppose instead you wanted to subtract the mean
value from each group. To do this, create a demeaning function and pass it
to transform:
In [231]: def demean(arr):
.....: return arr - arr.mean()
In [232]: demeaned = people.groupby(key).transform(demean)
In [233]: demeaned
Out[233]:
a b c d e
Jim -0.381154 1.152125 0.447807 -0.834043 0.891190
Joe 1.089221 -0.232534 1.322612 1.113271 1.381226
Steve 0.381154 -1.152125 -0.447807 0.834043 -0.891190
Travis -0.631512 0.232534 -1.322612 -0.976402 -0.832448
Wes -0.457709 NaN NaN -0.136869 -0.548778You can check that demeaned now
has zero group means:
In [234]: demeaned.groupby(key).mean()
Out[234]:
a b c d e
one -0.152570 0.000000e+00 0 -0.045623 -0.182926
two 0.228855 -1.110223e-16 0 0.068435 0.274389As you’ll see in the next section, group demeaning can be achieved
using apply also.
Like aggregate,
transform is a more specialized function
having rigid requirements: the passed function must either produce a
scalar value to be broadcasted (like np.mean) or a transformed array of the same
size. The most general purpose GroupBy method is apply, which is the subject of the rest of
this section. As in Figure 9-1, apply splits the object being manipulated into
pieces, invokes the passed function on each piece, then attempts to
concatenate the pieces together.
Returning to the tipping data set above, suppose you wanted to
select the top five tip_pct values by
group. First, it’s straightforward to write a function that selects the
rows with the largest values in a particular column:
In [235]: def top(df, n=5, column='tip_pct'):
.....: return df.sort_index(by=column)[-n:]
In [236]: top(tips, n=6)
Out[236]:
total_bill tip sex smoker day time size tip_pct
109 14.31 4.00 Female Yes Sat Dinner 2 0.279525
183 23.17 6.50 Male Yes Sun Dinner 4 0.280535
232 11.61 3.39 Male No Sat Dinner 2 0.291990
67 3.07 1.00 Female Yes Sat Dinner 1 0.325733
178 9.60 4.00 Female Yes Sun Dinner 2 0.416667
172 7.25 5.15 Male Yes Sun Dinner 2 0.710345Now, if we group by smoker,
say, and call apply with this
function, we get the following:
In [237]: tips.groupby('smoker').apply(top)
Out[237]:
total_bill tip sex smoker day time size tip_pct
smoker
No 88 24.71 5.85 Male No Thur Lunch 2 0.236746
185 20.69 5.00 Male No Sun Dinner 5 0.241663
51 10.29 2.60 Female No Sun Dinner 2 0.252672
149 7.51 2.00 Male No Thur Lunch 2 0.266312
232 11.61 3.39 Male No Sat Dinner 2 0.291990
Yes 109 14.31 4.00 Female Yes Sat Dinner 2 0.279525
183 23.17 6.50 Male Yes Sun Dinner 4 0.280535
67 3.07 1.00 Female Yes Sat Dinner 1 0.325733
178 9.60 4.00 Female Yes Sun Dinner 2 0.416667
172 7.25 5.15 Male Yes Sun Dinner 2 0.710345What has happened here? The top function is called
on each piece of the DataFrame, then the results are glued together
using pandas.concat, labeling
the pieces with the group names. The result therefore has a hierarchical
index whose inner level contains index values from the original
DataFrame.
If you pass a function to apply
that takes other arguments or keywords, you can pass these after the
function:
In [238]: tips.groupby(['smoker', 'day']).apply(top, n=1, column='total_bill')
Out[238]:
total_bill tip sex smoker day time size
smoker day
No Fri 94 22.75 3.25 Female No Fri Dinner 2
Sat 212 48.33 9.00 Male No Sat Dinner 4
Sun 156 48.17 5.00 Male No Sun Dinner 6
Thur 142 41.19 5.00 Male No Thur Lunch 5
Yes Fri 95 40.17 4.73 Male Yes Fri Dinner 4
Sat 170 50.81 10.00 Male Yes Sat Dinner 3
Sun 182 45.35 3.50 Male Yes Sun Dinner 3
Thur 197 43.11 5.00 Female Yes Thur Lunch 4
tip_pct
smoker day
No Fri 94 0.142857
Sat 212 0.186220
Sun 156 0.103799
Thur 142 0.121389
Yes Fri 95 0.117750
Sat 170 0.196812
Sun 182 0.077178
Thur 197 0.115982Note
Beyond these basic usage mechanics, getting the most out of
apply is largely a matter of
creativity. What occurs inside the function passed is up to you; it
only needs to return a pandas object or a scalar value. The rest of
this chapter will mainly consist of examples showing you how to solve
various problems using groupby.
You may recall above I called describe on a GroupBy
object:
In [239]: result = tips.groupby('smoker')['tip_pct'].describe()
In [240]: result
Out[240]:
smoker
No count 151.000000
mean 0.159328
std 0.039910
...
Yes 50% 0.153846
75% 0.195059
max 0.710345
Length: 16, dtype: float64
In [241]: result.unstack('smoker')
Out[241]:
smoker No Yes
count 151.000000 93.000000
mean 0.159328 0.163196
std 0.039910 0.085119
min 0.056797 0.035638
25% 0.136906 0.106771
50% 0.155625 0.153846
75% 0.185014 0.195059
max 0.291990 0.710345Inside GroupBy, when you invoke a method like describe, it is actually just a shortcut
for:
f = lambda x: x.describe() grouped.apply(f)
In the examples above, you see that the resulting object
has a hierarchical index formed from the group keys along with the
indexes of each piece of the original object. This can be disabled by
passing group_keys=False to
groupby:
In [242]: tips.groupby('smoker', group_keys=False).apply(top)
Out[242]:
total_bill tip sex smoker day time size tip_pct
88 24.71 5.85 Male No Thur Lunch 2 0.236746
185 20.69 5.00 Male No Sun Dinner 5 0.241663
51 10.29 2.60 Female No Sun Dinner 2 0.252672
149 7.51 2.00 Male No Thur Lunch 2 0.266312
232 11.61 3.39 Male No Sat Dinner 2 0.291990
109 14.31 4.00 Female Yes Sat Dinner 2 0.279525
183 23.17 6.50 Male Yes Sun Dinner 4 0.280535
67 3.07 1.00 Female Yes Sat Dinner 1 0.325733
178 9.60 4.00 Female Yes Sun Dinner 2 0.416667
172 7.25 5.15 Male Yes Sun Dinner 2 0.710345As you may recall from Chapter 7,
pandas has some tools, in particular cut and qcut, for slicing data
up into buckets with bins of your choosing or by sample quantiles.
Combining these functions with groupby, it becomes very simple to perform
bucket or quantile analysis on a data set. Consider a simple random data
set and an equal-length bucket categorization using cut:
In [243]: frame = DataFrame({'data1': np.random.randn(1000),
.....: 'data2': np.random.randn(1000)})
In [244]: factor = pd.cut(frame.data1, 4)
In [245]: factor[:10]
Out[245]:
(-1.23, 0.489]
(-2.956, -1.23]
(-1.23, 0.489]
(0.489, 2.208]
(-1.23, 0.489]
(0.489, 2.208]
(-1.23, 0.489]
(-1.23, 0.489]
(0.489, 2.208]
(0.489, 2.208]
Levels (4): Index(['(-2.956, -1.23]', '(-1.23, 0.489]',
'(0.489, 2.208]', '(2.208, 3.928]'], dtype=object)The Factor object returned
by cut can be passed directly to
groupby. So we could compute a set of
statistics for the data2 column like
so:
def get_stats(group):
return {'min': group.min(), 'max': group.max(),
'count': group.count(), 'mean': group.mean()}
grouped = frame.data2.groupby(factor)
grouped.apply(get_stats).unstack()These were equal-length buckets; to compute equal-size buckets
based on sample quantiles, use qcut. I’ll pass
labels=False to just get quantile
numbers.
# Return quantile numbers grouping = pd.qcut(frame.data1, 10, labels=False) grouped = frame.data2.groupby(grouping) grouped.apply(get_stats).unstack()
When cleaning up missing data, in some cases you will
filter out data observations using dropna, but in others you may want to impute
(fill in) the NA values using a fixed value or some value derived from
the data. fillna is the right
tool to use; for example here I fill in NA values with the mean:
In [248]: s = Series(np.random.randn(6)) In [249]: s[::2] = np.nan In [250]: s Out[250]: 0 NaN 1 -0.125921 2 NaN 3 -0.884475 4 NaN 5 0.227290 dtype: float64 In [251]: s.fillna(s.mean()) Out[251]: 0 -0.261035 1 -0.125921 2 -0.261035 3 -0.884475 4 -0.261035 5 0.227290 dtype: float64
Suppose you need the fill value to vary by group. As you may
guess, you need only group the data and use apply with a function
that calls fillna on each data
chunk. Here is some sample data on some US states divided into eastern
and western states:
In [252]: states = ['Ohio', 'New York', 'Vermont', 'Florida', .....: 'Oregon', 'Nevada', 'California', 'Idaho'] In [253]: group_key = ['East'] * 4 + ['West'] * 4 In [254]: data = Series(np.random.randn(8), index=states) In [255]: data[['Vermont', 'Nevada', 'Idaho']] = np.nan In [256]: data Out[256]: Ohio 0.922264 New York -2.153545 Vermont NaN Florida -0.375842 Oregon 0.329939 Nevada NaN California 1.105913 Idaho NaN dtype: float64 In [257]: data.groupby(group_key).mean() Out[257]: East -0.535707 West 0.717926 dtype: float64
We can fill the NA values using the group means like so:
In [258]: fill_mean = lambda g: g.fillna(g.mean()) In [259]: data.groupby(group_key).apply(fill_mean) Out[259]: Ohio 0.922264 New York -2.153545 Vermont -0.535707 Florida -0.375842 Oregon 0.329939 Nevada 0.717926 California 1.105913 Idaho 0.717926 dtype: float64
In another case, you might have pre-defined fill values in your
code that vary by group. Since the groups have a name attribute set internally, we can use
that:
fill_values = {'East': 0.5, 'West': -1}
fill_func = lambda g: g.fillna(fill_values[g.name])
data.groupby(group_key).apply(fill_func)Suppose you wanted to draw a random sample (with or
without replacement) from a large dataset for Monte Carlo simulation
purposes or some other application. There are a number of ways to
perform the “draws”; some are much more efficient than others. One way
is to select the first K elements of
np.random.permutation(N), where
N is the size of your complete
dataset and K the desired sample
size. As a more fun example, here’s a way to construct a deck of
English-style playing cards:
# Hearts, Spades, Clubs, Diamonds
suits = ['H', 'S', 'C', 'D']
card_val = (range(1, 11) + [10] * 3) * 4
base_names = ['A'] + range(2, 11) + ['J', 'K', 'Q']
cards = []
for suit in ['H', 'S', 'C', 'D']:
cards.extend(str(num) + suit for num in base_names)
deck = Series(card_val, index=cards)So now we have a Series of length 52 whose index contains card names and values are the ones used in blackjack and other games (to keep things simple, I just let the ace be 1):
In [263]: deck[:13] Out[263]: AH 1 2H 2 3H 3 ... JH 10 KH 10 QH 10 Length: 13, dtype: int64
Now, based on what I said above, drawing a hand of 5 cards from the desk could be written as:
In [264]: def draw(deck, n=5): .....: return deck.take(np.random.permutation(len(deck))[:n]) In [265]: draw(deck) Out[265]: AD 1 8C 8 5H 5 KC 10 2C 2 dtype: int64
Suppose you wanted two random cards from each suit. Because the
suit is the last character of each card name, we can group based on this
and use apply:
In [266]: get_suit = lambda card: card[-1] # last letter is suit In [267]: deck.groupby(get_suit).apply(draw, n=2) Out[267]: C 2C 2 3C 3 D KD 10 8D 8 H KH 10 3H 3 S 2S 2 4S 4 dtype: int64 # alternatively In [268]: deck.groupby(get_suit, group_keys=False).apply(draw, n=2) Out[268]: KC 10 JC 10 AD 1 5D 5 5H 5 6H 6 7S 7 KS 10 dtype: int64
Under the split-apply-combine paradigm of groupby, operations between columns in a
DataFrame or two Series, such a group weighted average, become a routine
affair. As an example, take this dataset containing group keys, values,
and some weights:
In [269]: df = DataFrame({'category': ['a', 'a', 'a', 'a', 'b', 'b', 'b', 'b'],
.....: 'data': np.random.randn(8),
.....: 'weights': np.random.rand(8)})
In [270]: df
Out[270]:
category data weights
0 a 1.561587 0.957515
1 a 1.219984 0.347267
2 a -0.482239 0.581362
3 a 0.315667 0.217091
4 b -0.047852 0.894406
5 b -0.454145 0.918564
6 b -0.556774 0.277825
7 b 0.253321 0.955905The group weighted average by category would then be:
In [271]: grouped = df.groupby('category')
In [272]: get_wavg = lambda g: np.average(g['data'], weights=g['weights'])
In [273]: grouped.apply(get_wavg)
Out[273]:
category
a 0.811643
b -0.122262
dtype: float64As a less trivial example, consider a data set from Yahoo! Finance
containing end of day prices for a few stocks and the S&P 500 index
(the SPX ticker):
In [274]: close_px = pd.read_csv('ch09/stock_px.csv', parse_dates=True, index_col=0)
In [275]: close_px.info()
<class 'pandas.core.frame.DataFrame'>
DatetimeIndex: 2214 entries, 2003-01-02 00:00:00 to 2011-10-14 00:00:00
Data columns (total 4 columns):
AAPL 2214 non-null float64
MSFT 2214 non-null float64
XOM 2214 non-null float64
SPX 2214 non-null float64
dtypes: float64(4)
In [276]: close_px[-4:]
Out[276]:
AAPL MSFT XOM SPX
2011-10-11 400.29 27.00 76.27 1195.54
2011-10-12 402.19 26.96 77.16 1207.25
2011-10-13 408.43 27.18 76.37 1203.66
2011-10-14 422.00 27.27 78.11 1224.58One task of interest might be to compute a DataFrame consisting of
the yearly correlations of daily returns (computed from percent changes)
with SPX. Here is one way to do
it:
In [277]: rets = close_px.pct_change().dropna()
In [278]: spx_corr = lambda x: x.corrwith(x['SPX'])
In [279]: by_year = rets.groupby(lambda x: x.year)
In [280]: by_year.apply(spx_corr)
Out[280]:
AAPL MSFT XOM SPX
2003 0.541124 0.745174 0.661265 1
2004 0.374283 0.588531 0.557742 1
2005 0.467540 0.562374 0.631010 1
2006 0.428267 0.406126 0.518514 1
2007 0.508118 0.658770 0.786264 1
2008 0.681434 0.804626 0.828303 1
2009 0.707103 0.654902 0.797921 1
2010 0.710105 0.730118 0.839057 1
2011 0.691931 0.800996 0.859975 1There is, of course, nothing to stop you from computing inter-column correlations:
# Annual correlation of Apple with Microsoft In [281]: by_year.apply(lambda g: g['AAPL'].corr(g['MSFT'])) Out[281]: 2003 0.480868 2004 0.259024 2005 0.300093 2006 0.161735 2007 0.417738 2008 0.611901 2009 0.432738 2010 0.571946 2011 0.581987 dtype: float64
In the same vein as the previous example, you can use
groupby to perform more complex
group-wise statistical analysis, as long as the function returns a
pandas object or scalar value. For example, I can define the following
regress function (using
the statsmodels econometrics library)
which executes an ordinary least squares (OLS) regression on each chunk
of data:
import statsmodels.api as sm def regress(data, yvar, xvars): Y = data[yvar] X = data[xvars] X['intercept'] = 1. result = sm.OLS(Y, X).fit() return result.params
Now, to run a yearly linear regression of AAPL on SPX
returns, I execute:
In [283]: by_year.apply(regress, 'AAPL', ['SPX'])
Out[283]:
SPX intercept
2003 1.195406 0.000710
2004 1.363463 0.004201
2005 1.766415 0.003246
2006 1.645496 0.000080
2007 1.198761 0.003438
2008 0.968016 -0.001110
2009 0.879103 0.002954
2010 1.052608 0.001261
2011 0.806605 0.001514A pivot table is a data summarization
tool frequently found in spreadsheet programs and other data analysis
software. It aggregates a table of data by one or more keys, arranging the
data in a rectangle with some of the group keys along the rows and some
along the columns. Pivot tables in Python with pandas are made possible
using the groupby facility described in
this chapter combined with reshape operations utilizing hierarchical
indexing. DataFrame has a pivot_table
method, and additionally there is a top-level pandas.pivot_table function. In addition to
providing a convenience interface to groupby, pivot_table also can add partial totals, also
known as margins.
Returning to the tipping data set, suppose I wanted to compute a
table of group means (the default pivot_table aggregation
type) arranged by sex and smoker on the rows:
In [284]: tips.pivot_table(index=['sex', 'smoker'])
Out[284]:
size tip tip_pct total_bill
sex smoker
Female No 2.592593 2.773519 0.156921 18.105185
Yes 2.242424 2.931515 0.182150 17.977879
Male No 2.711340 3.113402 0.160669 19.791237
Yes 2.500000 3.051167 0.152771 22.284500This could have been easily produced using groupby. Now, suppose we want to aggregate only
tip_pct and size, and additionally group by day. I’ll put smoker in the table columns and day in the rows:
In [285]: tips.pivot_table(['tip_pct', 'size'], index=['sex', 'day'],
.....: columns='smoker')
Out[285]:
tip_pct size
smoker No Yes No Yes
sex day
Female Fri 0.165296 0.209129 2.500000 2.000000
Sat 0.147993 0.163817 2.307692 2.200000
Sun 0.165710 0.237075 3.071429 2.500000
Thur 0.155971 0.163073 2.480000 2.428571
Male Fri 0.138005 0.144730 2.000000 2.125000
Sat 0.162132 0.139067 2.656250 2.629630
Sun 0.158291 0.173964 2.883721 2.600000
Thur 0.165706 0.164417 2.500000 2.300000This table could be augmented to include partial totals by passing
margins=True. This has the effect of
adding All row and column labels, with
corresponding values being the group statistics for all the data within a
single tier. In this below example, the All values are means without taking into account
smoker vs. non-smoker (the All columns)
or any of the two levels of grouping on the rows (the All row):
In [286]: tips.pivot_table(['tip_pct', 'size'], index=['sex', 'day'],
.....: columns='smoker', margins=True)
Out[286]:
tip_pct size
smoker No Yes All No Yes All
sex day
Female Fri 0.165296 0.209129 0.199388 2.500000 2.000000 2.111111
Sat 0.147993 0.163817 0.156470 2.307692 2.200000 2.250000
Sun 0.165710 0.237075 0.181569 3.071429 2.500000 2.944444
Thur 0.155971 0.163073 0.157525 2.480000 2.428571 2.468750
Male Fri 0.138005 0.144730 0.143385 2.000000 2.125000 2.100000
Sat 0.162132 0.139067 0.151577 2.656250 2.629630 2.644068
Sun 0.158291 0.173964 0.162344 2.883721 2.600000 2.810345
Thur 0.165706 0.164417 0.165276 2.500000 2.300000 2.433333
All 0.159328 0.163196 0.160803 2.668874 2.408602 2.569672To use a different aggregation function, pass it to aggfunc. For example, 'count' or len will give you a cross-tabulation (count or
frequency) of group sizes:
In [287]: tips.pivot_table('tip_pct', index=['sex', 'smoker'], columns='day',
.....: aggfunc=len, margins=True)
Out[287]:
day Fri Sat Sun Thur All
sex smoker
Female No 2 13 14 25 54
Yes 7 15 4 7 33
Male No 2 32 43 20 97
Yes 8 27 15 10 60
All 19 87 76 62 244If some combinations are empty (or otherwise NA), you may wish to
pass a fill_value:
In [288]: tips.pivot_table('size', index=['time', 'sex', 'smoker'],
.....: columns='day', aggfunc='sum', fill_value=0)
Out[288]:
day Fri Sat Sun Thur
time sex smoker
Dinner Female No 2 30 43 2
Yes 8 33 10 0
Male No 4 85 124 0
Yes 12 71 39 0
Lunch Female No 3 0 0 60
Yes 6 0 0 17
Male No 0 0 0 50
Yes 5 0 0 23See Table 9-2 for a summary of pivot_table methods.
Table 9-2. pivot_table options
A cross-tabulation (or crosstab for short) is a special case of a pivot table that computes group frequencies. Here is a canonical example taken from the Wikipedia page on cross-tabulation:
In [292]: data Out[292]: Sample Gender Handedness 0 1 Female Right-handed 1 2 Male Left-handed 2 3 Female Right-handed 3 4 Male Right-handed 4 5 Male Left-handed 5 6 Male Right-handed 6 7 Female Right-handed 7 8 Female Left-handed 8 9 Male Right-handed 9 10 Female Right-handed
As part of some survey analysis, we might want to summarize this
data by gender and handedness. You could use pivot_table to do this, but the pandas.crosstab function is very
convenient:
In [293]: pd.crosstab(data.Gender, data.Handedness, margins=True) Out[293]: Handedness Left-handed Right-handed All Gender Female 1 4 5 Male 2 3 5 All 3 7 10
The first two arguments to crosstab can each either be an array or Series
or a list of arrays. As in the tips data:
In [294]: pd.crosstab([tips.time, tips.day], tips.smoker, margins=True)
Out[294]:
smoker No Yes All
time day
Dinner Fri 3 9 12
Sat 45 42 87
Sun 57 19 76
Thur 1 0 1
Lunch Fri 1 6 7
Thur 44 17 61
All 151 93 244The US Federal Election Commission publishes data on
contributions to political campaigns. This includes contributor names,
occupation and employer, address, and contribution amount. An interesting
dataset is from the 2012 US presidential election (http://www.fec.gov/disclosurep/PDownload.do). As of this
writing (June 2012), the full dataset for all states is a 150 megabyte CSV
file P00000001-ALL.csv, which can be
loaded with pandas.read_csv:
In [13]: fec = pd.read_csv('ch09/P00000001-ALL.csv')In [14]: fec.info() Out[14]: <class 'pandas.core.frame.DataFrame'> Int64Index: 1001731 entries, 0 to 1001730 Data columns: cmte_id 1001731 non-null values cand_id 1001731 non-null values cand_nm 1001731 non-null values contbr_nm 1001731 non-null values contbr_city 1001716 non-null values contbr_st 1001727 non-null values contbr_zip 1001620 non-null values contbr_employer 994314 non-null values contbr_occupation 994433 non-null values contb_receipt_amt 1001731 non-null values contb_receipt_dt 1001731 non-null values receipt_desc 14166 non-null values memo_cd 92482 non-null values memo_text 97770 non-null values form_tp 1001731 non-null values file_num 1001731 non-null values dtypes: float64(1), int64(1), object(14)
A sample record in the DataFrame looks like this:
In [15]: fec.ix[123456] Out[15]: cmte_id C00431445 cand_id P80003338 cand_nm Obama, Barack contbr_nm ELLMAN, IRA contbr_city TEMPE contbr_st AZ contbr_zip 852816719 contbr_employer ARIZONA STATE UNIVERSITY contbr_occupation PROFESSOR contb_receipt_amt 50 contb_receipt_dt 01-DEC-11 receipt_desc NaN memo_cd NaN memo_text NaN form_tp SA17A file_num 772372 Name: 123456
You can probably think of many ways to start slicing and dicing this data to extract informative statistics about donors and patterns in the campaign contributions. I’ll spend the next several pages showing you a number of different analyses that apply techniques you have learned about so far.
You can see that there are no political party affiliations in the
data, so this would be useful to add. You can get a list of all the unique
political candidates using unique (note that NumPy
suppresses the quotes around the strings in the output):
In [16]: unique_cands = fec.cand_nm.unique()
In [17]: unique_cands
Out[17]:
array([Bachmann, Michelle, Romney, Mitt, Obama, Barack,
Roemer, Charles E. 'Buddy' III, Pawlenty, Timothy,
Johnson, Gary Earl, Paul, Ron, Santorum, Rick, Cain, Herman,
Gingrich, Newt, McCotter, Thaddeus G, Huntsman, Jon, Perry, Rick], dtype=object)
In [18]: unique_cands[2]
Out[18]: 'Obama, Barack'An easy way to indicate party affiliation is using a dict:[4]
parties = {'Bachmann, Michelle': 'Republican',
'Cain, Herman': 'Republican',
'Gingrich, Newt': 'Republican',
'Huntsman, Jon': 'Republican',
'Johnson, Gary Earl': 'Republican',
'McCotter, Thaddeus G': 'Republican',
'Obama, Barack': 'Democrat',
'Paul, Ron': 'Republican',
'Pawlenty, Timothy': 'Republican',
'Perry, Rick': 'Republican',
"Roemer, Charles E. 'Buddy' III": 'Republican',
'Romney, Mitt': 'Republican',
'Santorum, Rick': 'Republican'}Now, using this mapping and the map method on Series
objects, you can compute an array of political parties from the candidate
names:
In [20]: fec.cand_nm[123456:123461] Out[20]: 123456 Obama, Barack 123457 Obama, Barack 123458 Obama, Barack 123459 Obama, Barack 123460 Obama, Barack Name: cand_nm In [21]: fec.cand_nm[123456:123461].map(parties) Out[21]: 123456 Democrat 123457 Democrat 123458 Democrat 123459 Democrat 123460 Democrat Name: cand_nm # Add it as a column In [22]: fec['party'] = fec.cand_nm.map(parties) In [23]: fec['party'].value_counts() Out[23]: Democrat 593746 Republican 407985
A couple of data preparation points. First, this data includes both contributions and refunds (negative contribution amount):
In [24]: (fec.contb_receipt_amt > 0).value_counts() Out[24]: True 991475 False 10256
To simplify the analysis, I’ll restrict the data set to positive contributions:
In [25]: fec = fec[fec.contb_receipt_amt > 0]
Since Barack Obama and Mitt Romney are the main two candidates, I’ll also prepare a subset that just has contributions to their campaigns:
In [26]: fec_mrbo = fec[fec.cand_nm.isin(['Obama, Barack', 'Romney, Mitt'])]
Donations by occupation is another oft-studied statistic. For example, lawyers (attorneys) tend to donate more money to Democrats, while business executives tend to donate more to Republicans. You have no reason to believe me; you can see for yourself in the data. First, the total number of donations by occupation is easy:
In [27]: fec.contbr_occupation.value_counts()[:10] Out[27]: RETIRED 233990 INFORMATION REQUESTED 35107 ATTORNEY 34286 HOMEMAKER 29931 PHYSICIAN 23432 INFORMATION REQUESTED PER BEST EFFORTS 21138 ENGINEER 14334 TEACHER 13990 CONSULTANT 13273 PROFESSOR 12555
You will notice by looking at the occupations that many refer to
the same basic job type, or there are several variants of the same
thing. Here is a code snippet illustrates a technique for cleaning up a
few of them by mapping from one occupation to another; note the “trick”
of using dict.get to allow
occupations with no mapping to “pass through”:
occ_mapping = {
'INFORMATION REQUESTED PER BEST EFFORTS' : 'NOT PROVIDED',
'INFORMATION REQUESTED' : 'NOT PROVIDED',
'INFORMATION REQUESTED (BEST EFFORTS)' : 'NOT PROVIDED',
'C.E.O.': 'CEO'
}
# If no mapping provided, return x
f = lambda x: occ_mapping.get(x, x)
fec.contbr_occupation = fec.contbr_occupation.map(f)I’ll also do the same thing for employers:
emp_mapping = {
'INFORMATION REQUESTED PER BEST EFFORTS' : 'NOT PROVIDED',
'INFORMATION REQUESTED' : 'NOT PROVIDED',
'SELF' : 'SELF-EMPLOYED',
'SELF EMPLOYED' : 'SELF-EMPLOYED',
}
# If no mapping provided, return x
f = lambda x: emp_mapping.get(x, x)
fec.contbr_employer = fec.contbr_employer.map(f)Now, you can use pivot_table to
aggregate the data by party and occupation, then filter down to the
subset that donated at least $2 million overall:
In [34]: by_occupation = fec.pivot_table('contb_receipt_amt',
....: index='contbr_occupation',
....: columns='party', aggfunc='sum')
In [35]: over_2mm = by_occupation[by_occupation.sum(1) > 2000000]
In [36]: over_2mm
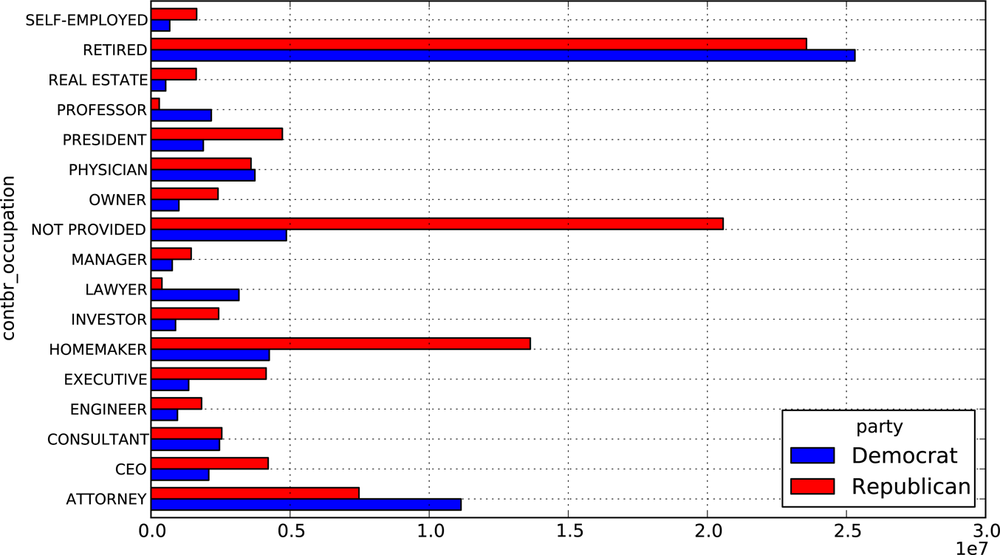
Out[36]:
party Democrat Republican
contbr_occupation
ATTORNEY 11141982.97 7477194.430000
CEO 2074974.79 4211040.520000
CONSULTANT 2459912.71 2544725.450000
ENGINEER 951525.55 1818373.700000
EXECUTIVE 1355161.05 4138850.090000
HOMEMAKER 4248875.80 13634275.780000
INVESTOR 884133.00 2431768.920000
LAWYER 3160478.87 391224.320000
MANAGER 762883.22 1444532.370000
NOT PROVIDED 4866973.96 20565473.010000
OWNER 1001567.36 2408286.920000
PHYSICIAN 3735124.94 3594320.240000
PRESIDENT 1878509.95 4720923.760000
PROFESSOR 2165071.08 296702.730000
REAL ESTATE 528902.09 1625902.250000
RETIRED 25305116.38 23561244.489999
SELF-EMPLOYED 672393.40 1640252.540000It can be easier to look at this data graphically as a bar plot
('barh' means horizontal bar plot,
see Figure 9-2):
In [38]: over_2mm.plot(kind='barh')
You might be interested in the top donor occupations or top
companies donating to Obama and Romney. To do this, you can group by
candidate name and use a variant of the top method from earlier
in the chapter:
def get_top_amounts(group, key, n=5):
totals = group.groupby(key)['contb_receipt_amt'].sum()
# Order totals by key in descending order
return totals.order(ascending=False)[-n:]Then aggregated by occupation and employer:
In [40]: grouped = fec_mrbo.groupby('cand_nm')
In [41]: grouped.apply(get_top_amounts, 'contbr_occupation', n=7)
Out[41]:
cand_nm contbr_occupation
Obama, Barack RETIRED 25305116.38
ATTORNEY 11141982.97
NOT PROVIDED 4866973.96
HOMEMAKER 4248875.80
PHYSICIAN 3735124.94
LAWYER 3160478.87
CONSULTANT 2459912.71
Romney, Mitt RETIRED 11508473.59
NOT PROVIDED 11396894.84
HOMEMAKER 8147446.22
ATTORNEY 5364718.82
PRESIDENT 2491244.89
EXECUTIVE 2300947.03
C.E.O. 1968386.11
Name: contb_receipt_amt
In [42]: grouped.apply(get_top_amounts, 'contbr_employer', n=10)
Out[42]:
cand_nm contbr_employer
Obama, Barack RETIRED 22694358.85
SELF-EMPLOYED 18626807.16
NOT EMPLOYED 8586308.70
NOT PROVIDED 5053480.37
HOMEMAKER 2605408.54
STUDENT 318831.45
VOLUNTEER 257104.00
MICROSOFT 215585.36
SIDLEY AUSTIN LLP 168254.00
REFUSED 149516.07
Romney, Mitt NOT PROVIDED 12059527.24
RETIRED 11506225.71
HOMEMAKER 8147196.22
SELF-EMPLOYED 7414115.22
STUDENT 496490.94
CREDIT SUISSE 281150.00
MORGAN STANLEY 267266.00
GOLDMAN SACH & CO. 238250.00
BARCLAYS CAPITAL 162750.00
H.I.G. CAPITAL 139500.00
Name: contb_receipt_amtA useful way to analyze this data is to use the cut function to
discretize the contributor amounts into buckets by contribution
size:
In [43]: bins = np.array([0, 1, 10, 100, 1000, 10000, 100000, 1000000, 10000000])
In [44]: labels = pd.cut(fec_mrbo.contb_receipt_amt, bins)
In [45]: labels
Out[45]:
Factor:contb_receipt_amt
array([(10, 100], (100, 1000], (100, 1000], ..., (1, 10], (10, 100],
(100, 1000]], dtype=object)
Levels (8): array([(0, 1], (1, 10], (10, 100], (100, 1000], (1000, 10000],
(10000, 100000], (100000, 1000000], (1000000, 10000000]], dtype=object)We can then group the data for Obama and Romney by name and bin label to get a histogram by donation size:
In [46]: grouped = fec_mrbo.groupby(['cand_nm', labels]) In [47]: grouped.size().unstack(0) Out[47]: cand_nm Obama, Barack Romney, Mitt contb_receipt_amt (0, 1] 493 77 (1, 10] 40070 3681 (10, 100] 372280 31853 (100, 1000] 153991 43357 (1000, 10000] 22284 26186 (10000, 100000] 2 1 (100000, 1000000] 3 NaN (1000000, 10000000] 4 NaN
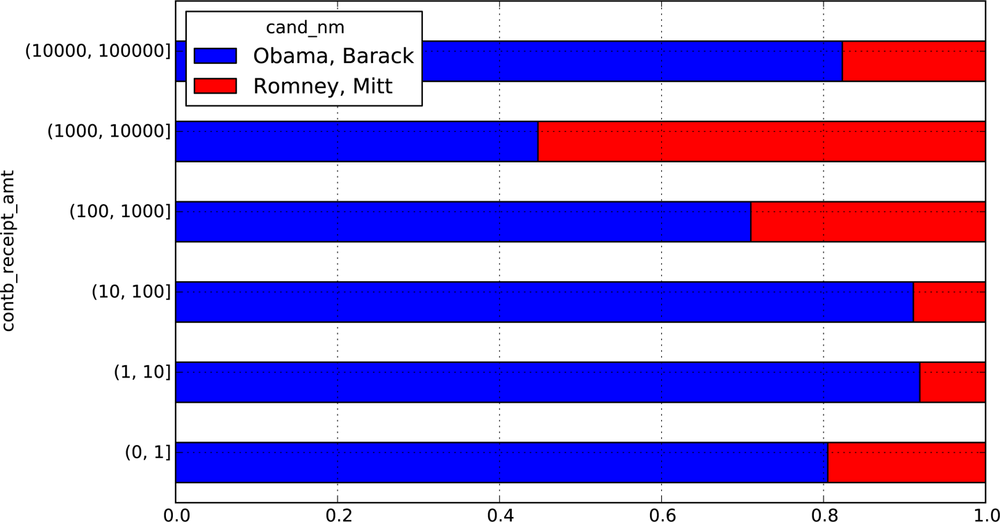
This data shows that Obama has received a significantly larger number of small donations than Romney. You can also sum the contribution amounts and normalize within buckets to visualize percentage of total donations of each size by candidate:
In [48]: bucket_sums = grouped.contb_receipt_amt.sum().unstack(0) In [49]: bucket_sums Out[49]: cand_nm Obama, Barack Romney, Mitt contb_receipt_amt (0, 1] 318.24 77.00 (1, 10] 337267.62 29819.66 (10, 100] 20288981.41 1987783.76 (100, 1000] 54798531.46 22363381.69 (1000, 10000] 51753705.67 63942145.42 (10000, 100000] 59100.00 12700.00 (100000, 1000000] 1490683.08 NaN (1000000, 10000000] 7148839.76 NaN In [50]: normed_sums = bucket_sums.div(bucket_sums.sum(axis=1), axis=0) In [51]: normed_sums Out[51]: cand_nm Obama, Barack Romney, Mitt contb_receipt_amt (0, 1] 0.805182 0.194818 (1, 10] 0.918767 0.081233 (10, 100] 0.910769 0.089231 (100, 1000] 0.710176 0.289824 (1000, 10000] 0.447326 0.552674 (10000, 100000] 0.823120 0.176880 (100000, 1000000] 1.000000 NaN (1000000, 10000000] 1.000000 NaN In [52]: normed_sums[:-2].plot(kind='barh', stacked=True)
I excluded the two largest bins as these are not donations by individuals. See Figure 9-3 for the resulting figure.
There are of course many refinements and improvements of this analysis. For example, you could aggregate donations by donor name and zip code to adjust for donors who gave many small amounts versus one or more large donations. I encourage you to download it and explore it yourself.
Aggregating the data by candidate and state is a routine affair:
In [53]: grouped = fec_mrbo.groupby(['cand_nm', 'contbr_st']) In [54]: totals = grouped.contb_receipt_amt.sum().unstack(0).fillna(0) In [55]: totals = totals[totals.sum(1) > 100000] In [56]: totals[:10] Out[56]: cand_nm Obama, Barack Romney, Mitt contbr_st AK 281840.15 86204.24 AL 543123.48 527303.51 AR 359247.28 105556.00 AZ 1506476.98 1888436.23 CA 23824984.24 11237636.60 CO 2132429.49 1506714.12 CT 2068291.26 3499475.45 DC 4373538.80 1025137.50 DE 336669.14 82712.00 FL 7318178.58 8338458.81
If you divide each row by the total contribution amount, you get the relative percentage of total donations by state for each candidate:
In [57]: percent = totals.div(totals.sum(1), axis=0) In [58]: percent[:10] Out[58]: cand_nm Obama, Barack Romney, Mitt contbr_st AK 0.765778 0.234222 AL 0.507390 0.492610 AR 0.772902 0.227098 AZ 0.443745 0.556255 CA 0.679498 0.320502 CO 0.585970 0.414030 CT 0.371476 0.628524 DC 0.810113 0.189887 DE 0.802776 0.197224 FL 0.467417 0.532583