Visualization is a powerful method to gain more insight about data and underlying processes. It is extensively used during the exploration phase to attain a better sense of how data is structured. Visual presentation of results is also an effective way to convey the key findings; people usually grasp nice and informative diagrams much easier than they grasp facts embedded in convoluted tabular form. Of course, this doesn’t mean that visualization should replace other approaches; the best effect is achieved by judiciously combining multiple ways to describe data and outcomes. The topic of visualization itself is very broad, so this chapter focuses on two aspects that are not commonly discussed elsewhere: how visualization may help in optimizing applications, and how to create dynamic dashboards for high-velocity data. You can find examples of other uses throughout this book.
Visualization should permeate data science processes and products. It cannot be treated as an isolated entity, let alone be reduced to fancy diagrams. In this respect, I frown upon any sort of rigid categorization and dislike embarking on templatizing what kind of visualization is appropriate in different scenarios. There are general guidelines about encoding of variables (see reference [1]) using various plot types, and these should be followed as much as possible. Nonetheless, the main message is that if visualization helps to make actionable insights from data, then it is useful.
Algorithms are the cornerstone of doing data science and dealing with Big Data. This is going to be demonstrated in the next case study. Algorithms specify the asymptotic behavior of your program (frequently denoted in Big-O notation), while technologies including parallel and distributed computing drive the constant factors. We will apply visualizations of various sorts to showcase these properties. It is important to emphasize that the notion of a program may symbolize many different types of computations. Computing features is one such type. A fast algorithm may enable your model to properly run in both the batch regime and online regime. For an example using interactive visualization to show the implementation of data structures and algorithms, you may want to visit the Data Structure Visualizations web site (see https://www.cs.usfca.edu/~galles/visualization ). Another interesting site for you to explore is Python Tutor, which is related to visualizing the execution of Python programs (visit https://pythontutor.com ). It also has links to similar sites for learning other popular languages, like JavaScript, C++, Java, etc.
Producing static graphs is totally different than trying to depict major characteristics of data in continual flux. This will be our topic in the second part of this chapter. Efficient dashboards allow real-time monitoring of processes by visualizing business metrics, so that operators may promptly act upon them.
Visualizing Temperature Data Case Study
The aim of this section is to illuminate the architecture of matplotlib, which is the most famous Python visualization framework (you should read the definitive guide at http://aosabook.org/en/matplotlib.html ). To make the topic more comprehensible, we will use a sample publicly available dataset from NOAA’s Global Historical Climatology Network - Daily (GHCN-Daily), Version 3. This sample dataset contains measurements from a single station for January 2010. It is available in the source code for this chapter, but you can download it directly from https://www1.ncdc.noaa.gov/pub/data/cdo/samples/GHCND_sample_csv.csv . For each day we will use the columns denoting the minimum and maximum temperature readings given in tenths of degree Celsius. The station’s latitude and longitude are expressed in the correspondingly named columns.
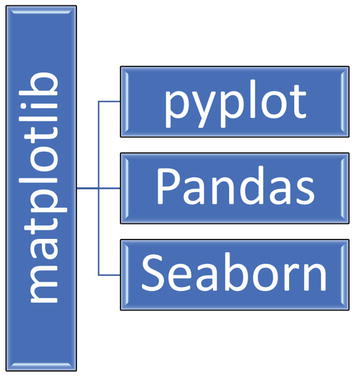
Some higher-level components that sit on top of matplotlib, which is further segregated into smaller units that you can reference
Showing Stations on a Map
Module for Showing Our Data Source on a Map.
First Part of the temp_visualization_demo.py Module
Figure 6-2 shows the generated interactive map with a blue diamond denoting our temperature sensor. You will also find the _map.html file in the current working directory. This is automatically created by mplleaflet.show() (it is possible to change the output file name).
Plotting Temperatures
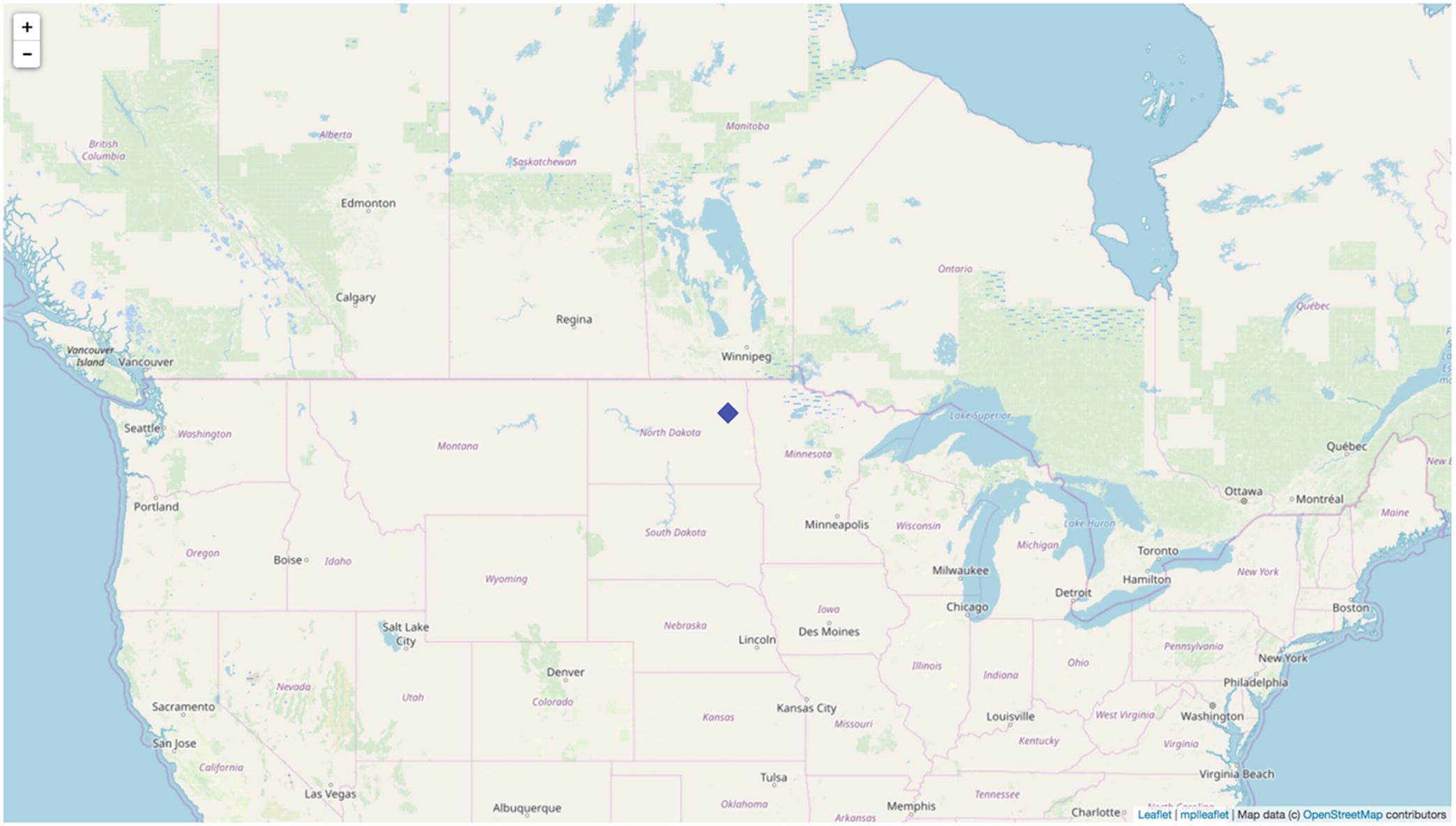
The interactive map with our temperature sensor (a quite remarkable result with just a couple of lines of code)
Second Part of the temp_visualization_demo.py Module
The Function plot_temps That Produces the Plot As Shown in Figure 6-3
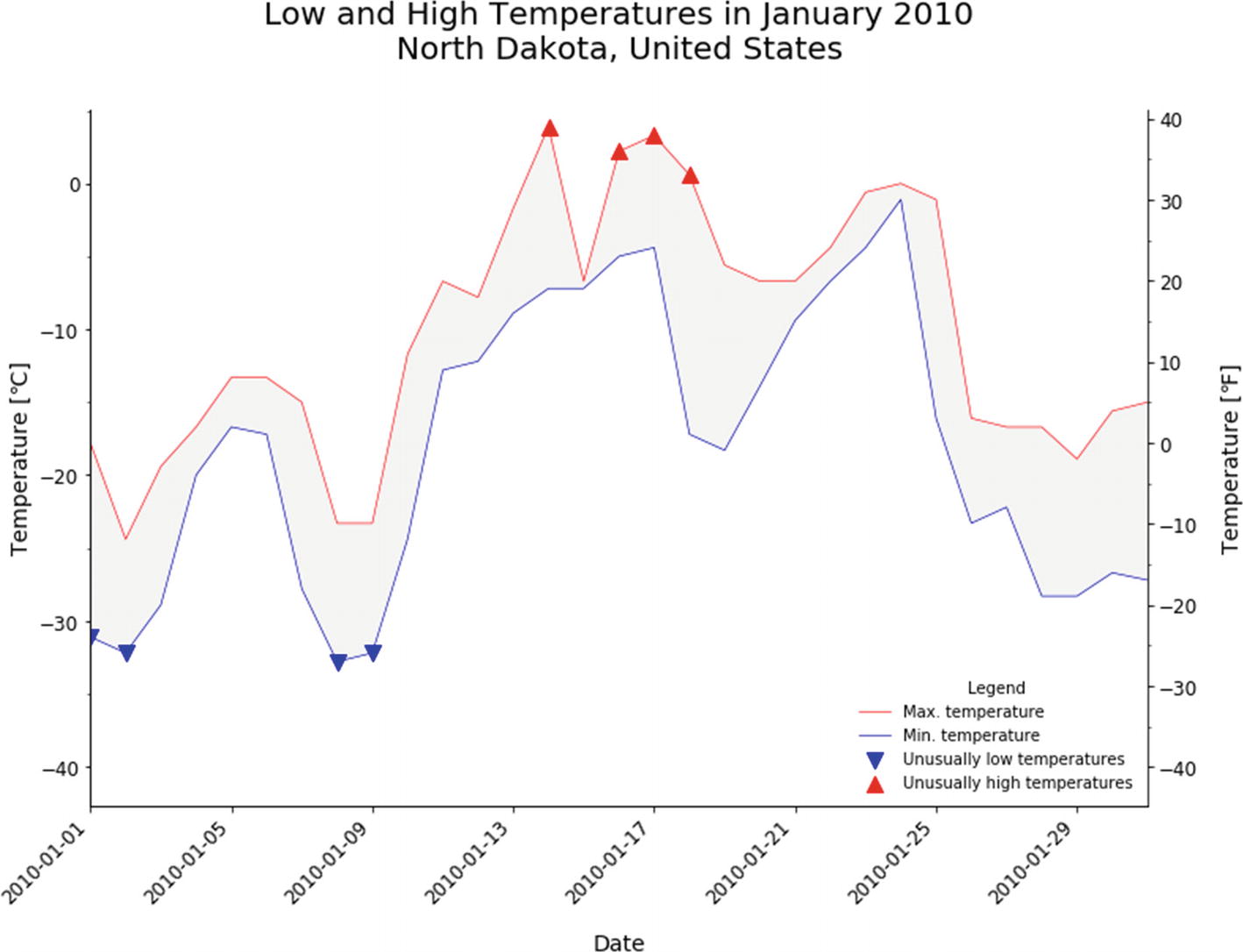
The plot of low and high temperatures with markers for extreme temperatures (defined via thresholds)
Closest Pair Case Study
Finding the closest pair is a well-known task from computational geometry with many applications
. We have a set of points in the plane given by their x and y coordinates, represented as P = {(x, y)| x, y ∈ ℤ}. We need to find the closest pair of points pi ≠ pj whose Euclidian distance is defined as  . Let us assume that the number of points n is in the range of [2, 107].
. Let us assume that the number of points n is in the range of [2, 107].
We will develop three core variants with different asymptotic behaviors: O(n2), O(nlog2n), and O(n log n). You can refresh your memory about asymptotic notation and algorithms by consulting references [2] through [4]. Afterward, we will bring down the constants to show how they affect the overall performance. Visualization will help us to illustrate the results more clearly. This is an example of using visualization to acquire deeper understanding of a program’s run-time trait.
Abstract Base Class Specifying the Shared API for All Subsequent Variants
Our next task is to create a small test harness that will measure execution times and create a proper visualization. It turns out that there are available software tools for this purpose: cProfile (read more details at https://docs.python.org/3/library/profile.html ) and SnakeViz (you may need to install it by executing conda install snakeviz from a command line). cProfile outputs data in a format that is directly consumable by SnakeViz.
Version 1.0
The Brute-Force Implementation of This Problem
Tip
Don’t underestimate the value of such a simple but straightforward variant like NaiveClosestPair. It can be a very useful baseline both for performance comparisons and for assurance of correctness. Namely, it can be used together with the point generator to produce many low-dimensional test cases.

The entry page of SnakeViz, where you can select the ncp_100_stats file for visualization
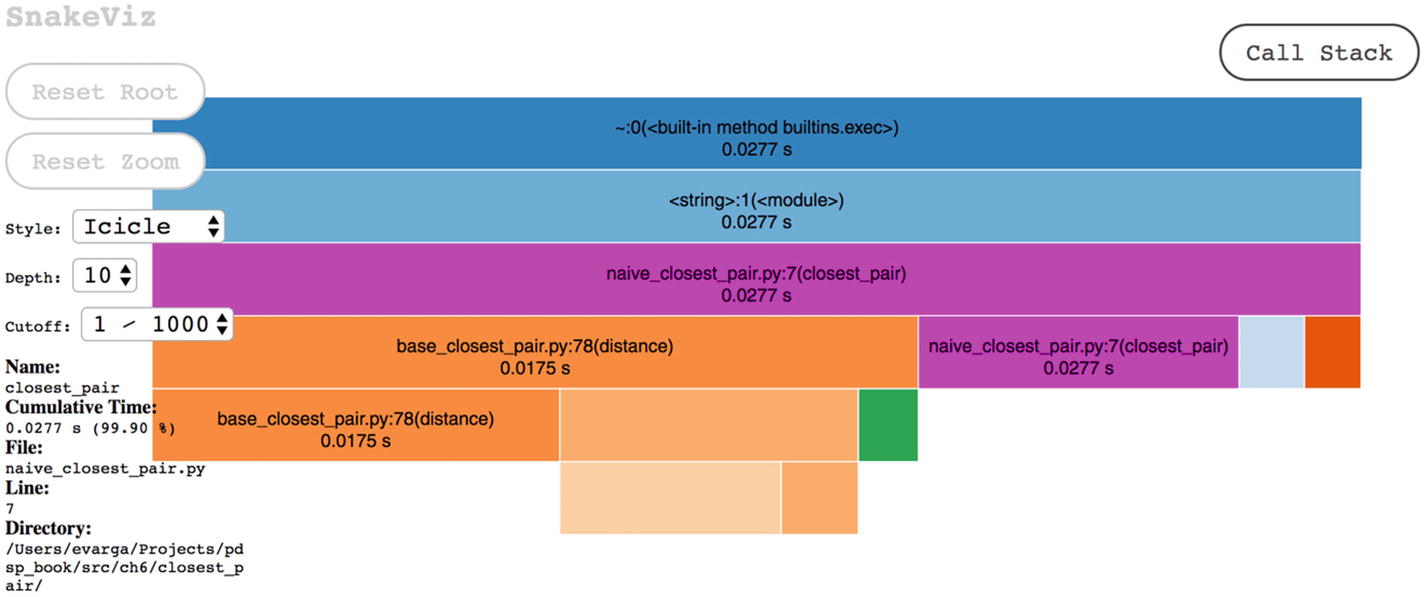
The diagram is interactive
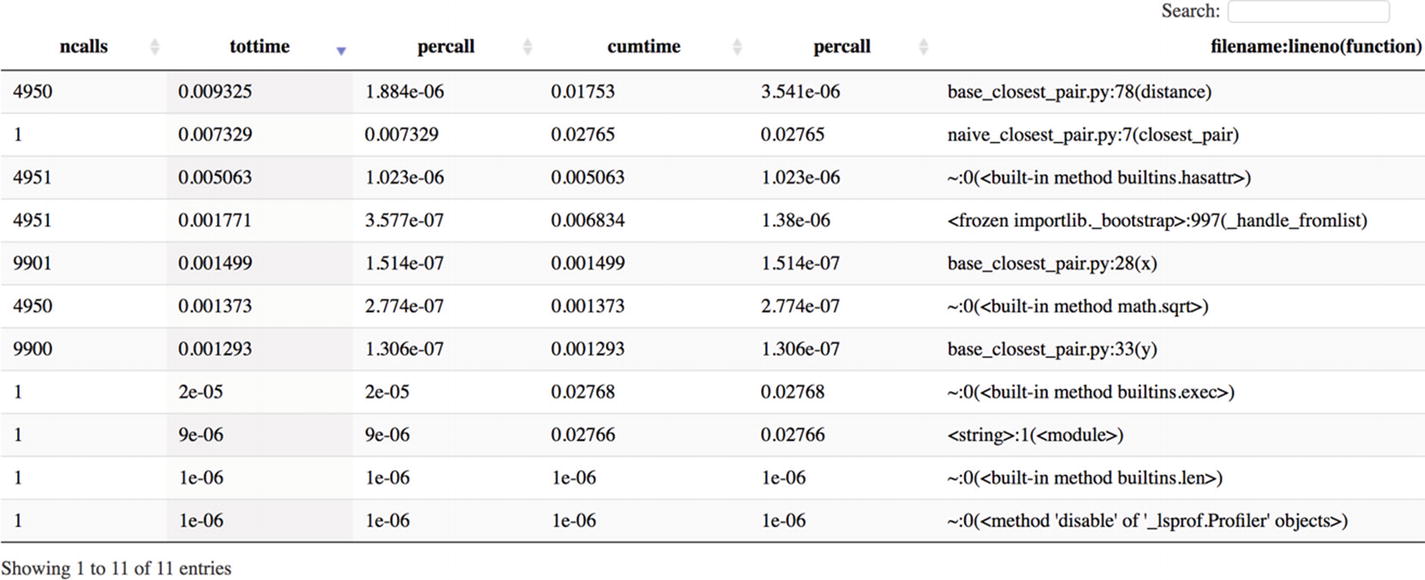
ncalls designates the number of calls made to a function (for example, 4950 = 100 × 99 / 2, which is the number of times distance was called), while tottime shows the total time spent inside a method
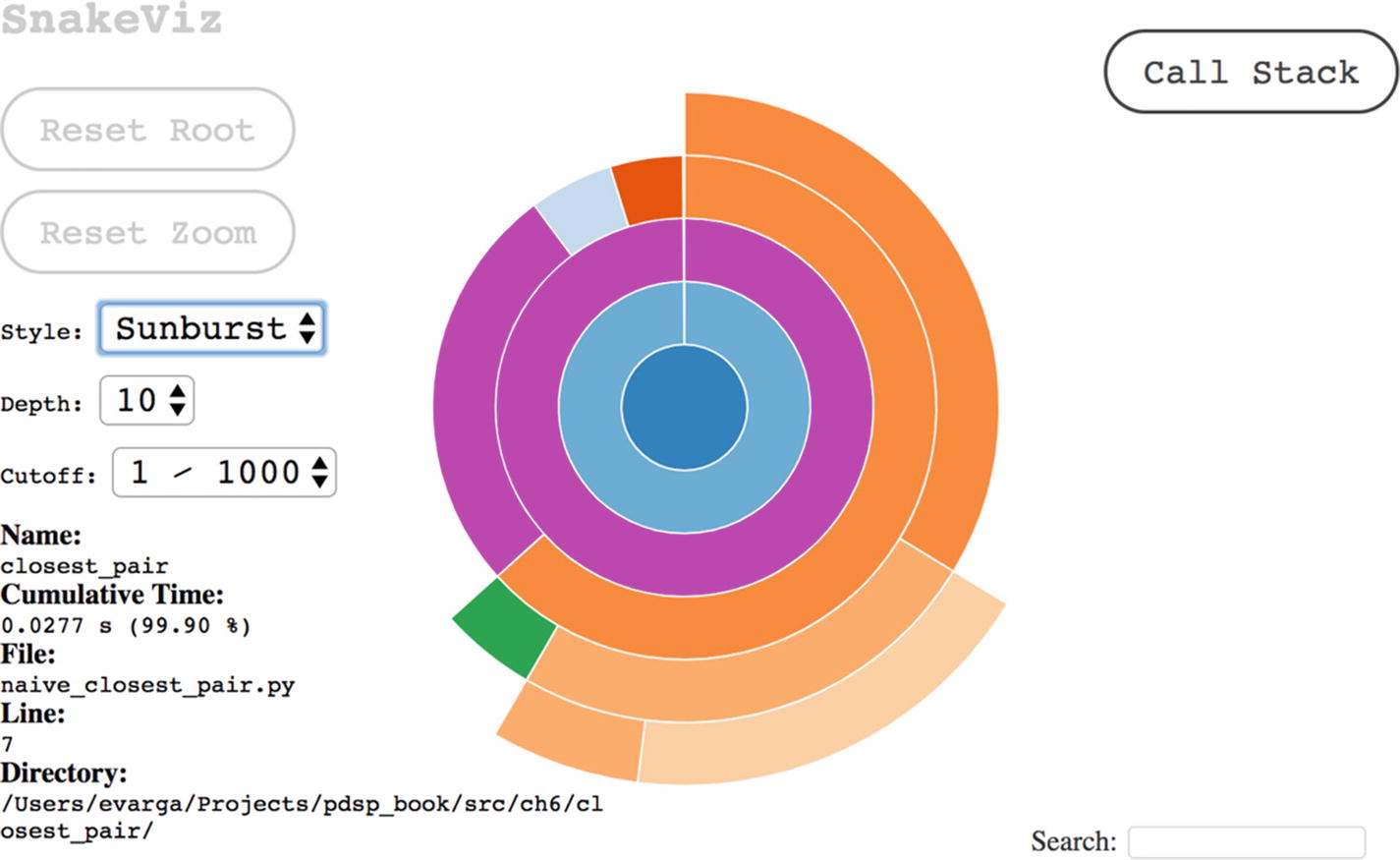
Another way to visualize the calling hierarchy, which may be useful to see the big picture
This will take much more to finish. Navigate back to the folder page of SnakeViz by visiting http://127.0.0.1:<port number>/snakeviz/results and selecting ncp_3000_stats. Sort the table by cumulative time in decreasing order and you will see that the running time is around 20 seconds (on my Mac it was 23.48s).1 The previous run with 100 points took only 0.0277s; a 30× increase of input size induced a roughly 900× upsurge of the running time. This is what O(n2) is all about. The exact times on your machine will be different, but the ratio will stay the same. Whatever you do to speed things up (for example, using Numba), you will remain at the same asymptotic level; in other words, the ratio of running times will mirror the quadratic dependence on input size. A pure Python code running a better algorithm will always outperform at one point an ultra-technologically boosted version that uses some slow algorithm.
Version 2.0
In this section we will lay out the O(nlog2n) variant of the algorithm that uses the divide-and-conquer technique . This technique divides the initial problem into multiple subproblems of the same type and solves them independently. If any subproblem is still complex, then it is further divided using the same procedure. The chain ends when you hit a base case (i.e., a problem that is trivial to solve). Afterward, the results of subproblems are combined until the final answer is composed. A representative example of this methodology is the merge sort.
- 1.
Split the points into two halves based upon their x coordinate. More formally, if we denote the cutting coordinate as x, then we want to produce two sets of points, L (stands for left) and R (stands for right), such that all points of L have xi ≤ x and all points of R have xi > x.
- 2.
Recursively treat groups L and R as separate subproblems and find their solutions as tuples (pL, qL, dL) and (pR, qR, dR), respectively. Take d = min(dL, dR) and remember from which group that minimum came.
- 3.
Check if there is any point in L and R whose distance is smaller than d. Select all points of L and R whose x coordinate belongs to interval [x – d, x + d]. Sort these middle (M) points by their y coordinate (the ordering doesn’t matter).
- 4.
Iterate over this sorted list, and for each point, calculate its distance to at most seven subsequent points and maintain the currently found best pair. Let it be the triple (pM, qM, dM).
- 5.
The final solution is the minimum among pairs of L, M, and R. Recall that in step 2 we already found the minimum between L and R, so we just need one extra comparison with M.
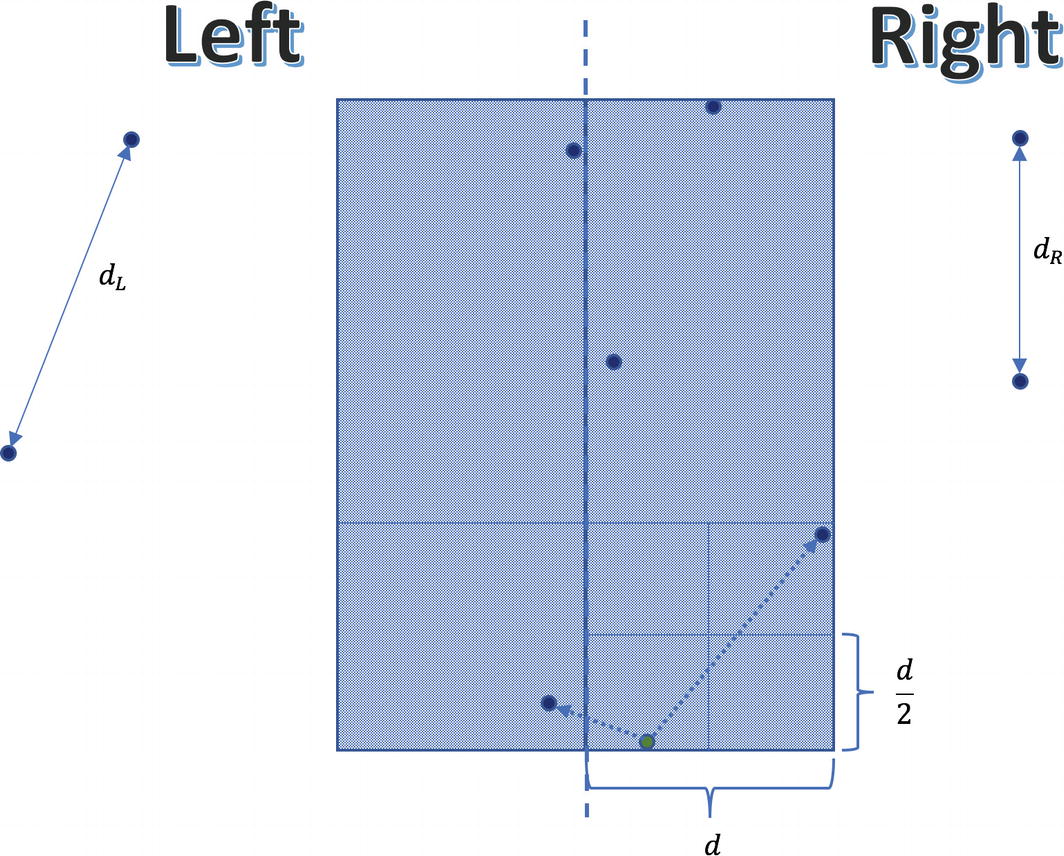
Notice that inside any block of size d×d there can be at most four points
Any of the OS kernel’s subblocks of size 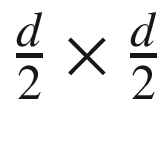 can only contain at most one point. In other words, if a square of dimensions d×d would contain five or more points, then some of its subblocks will surely host more than one point. Nonetheless, in this case we would reach a contradiction that d isn’t correctly computed. Furthermore, any two points in this strip whose distance is < d must be part of the same rectangle of size 2d×d. Consequently, they definitely will be compared, which guarantees correctness. Recall that this rectangle may encompass at most eight points (including the reference point). Here, we will find a shorter distance than d, because the distance between the lowest-right point and subsequent left point is small.
can only contain at most one point. In other words, if a square of dimensions d×d would contain five or more points, then some of its subblocks will surely host more than one point. Nonetheless, in this case we would reach a contradiction that d isn’t correctly computed. Furthermore, any two points in this strip whose distance is < d must be part of the same rectangle of size 2d×d. Consequently, they definitely will be compared, which guarantees correctness. Recall that this rectangle may encompass at most eight points (including the reference point). Here, we will find a shorter distance than d, because the distance between the lowest-right point and subsequent left point is small.
Analysis of the Running Time
Assume that we are given n points in the plane. In step 1 we can find the median point that splits the set into two equally sized subsets, in O(n) time using a randomized divide-and-conquer algorithm. Creating subsets L and R is another O(n) time operation. After solving both subproblems of size 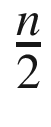 the algorithm selects points belonging to the middle strip in O(n) time. Afterward, it sorts these filtered points in O(n log n) time. Finally, it computes distances inside this strip in O(n) time (for each point it does some fixed amount of work). Remember that for any function f = kn, k ∈ ℕ it is true that f = O(n); that is, it is bounded by some linear function. Therefore, irrespectively of how many times we do O(n) operations in succession or whether we do 3n, 5n, or 7n operations, their aggregate effect counts as O(n). All in all, the overall running time is defined by the recurrent formula
the algorithm selects points belonging to the middle strip in O(n) time. Afterward, it sorts these filtered points in O(n log n) time. Finally, it computes distances inside this strip in O(n) time (for each point it does some fixed amount of work). Remember that for any function f = kn, k ∈ ℕ it is true that f = O(n); that is, it is bounded by some linear function. Therefore, irrespectively of how many times we do O(n) operations in succession or whether we do 3n, 5n, or 7n operations, their aggregate effect counts as O(n). All in all, the overall running time is defined by the recurrent formula 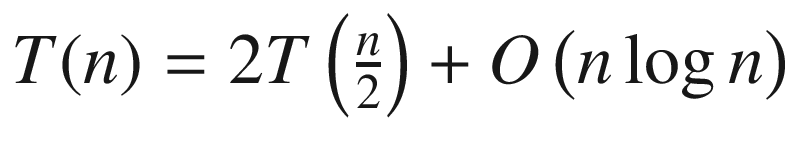 . We cannot directly apply the Master Theorem, although we may reuse its idea for analyzing our expression.
. We cannot directly apply the Master Theorem, although we may reuse its idea for analyzing our expression.
If we draw a recursion tree, then at some arbitrary level k, the tree’s contribution would be 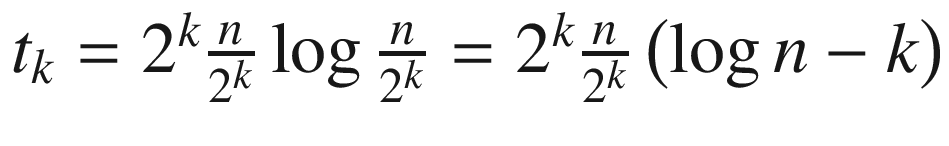 . The depth of the tree is logn, so by summing up all contributions from each level, we end up with
. The depth of the tree is logn, so by summing up all contributions from each level, we end up with 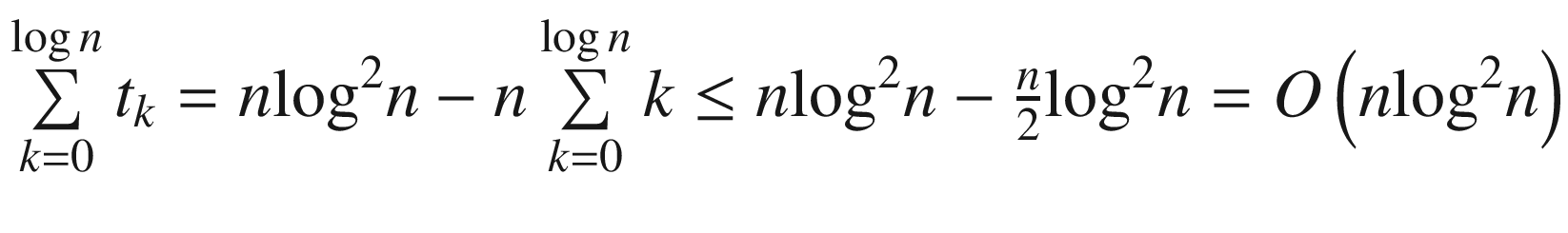 .
.
In step 3 we repeatedly sort points by their y coordinate, and this amounts to that extra logn multiplier. If we could eschew this sorting by only doing it once at the beginning, then we would lower the running time to O(n log n). This is evident from the new recurrence relation 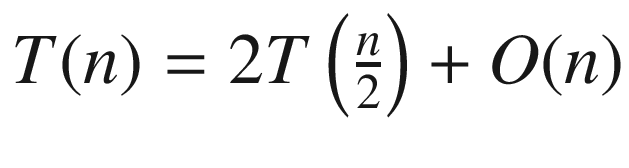 , since we may immediately apply the Master Theorem. Of course, we should be careful to preserve the y ordering of points during all sorts of splitting and filtering actions.
, since we may immediately apply the Master Theorem. Of course, we should be careful to preserve the y ordering of points during all sorts of splitting and filtering actions.
Version 3.0
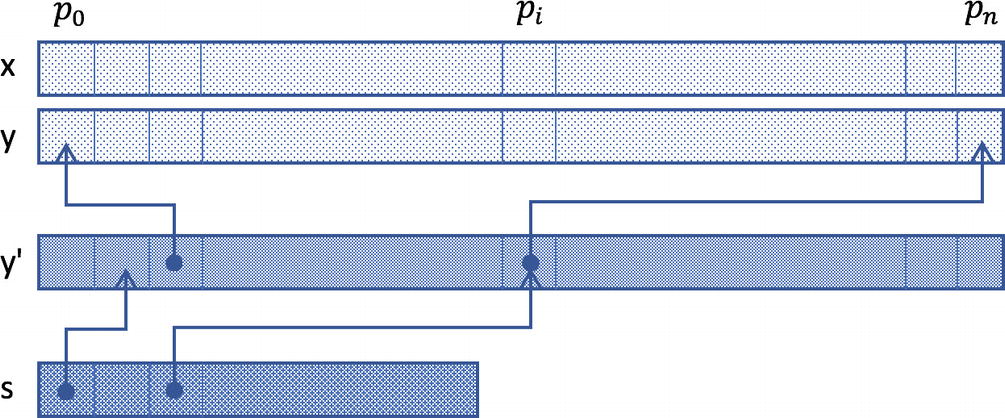
The array s denotes the ordered indices of points belonging to the middle strip
Implementation of the FastClosestPair Class
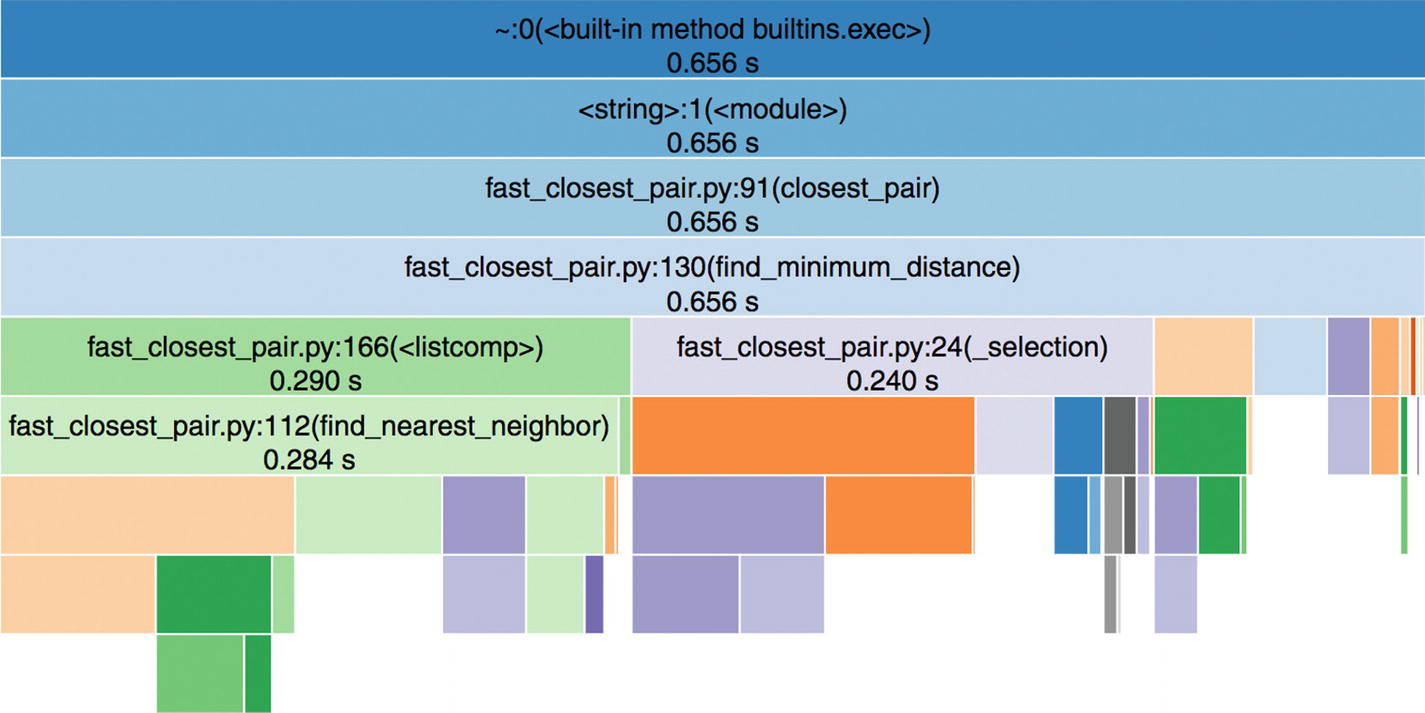
The picture only shows the Icicle-style diagram. Apparently, there are many additional details, since the code base is more involved.
Enquiry of Algorithms Evolution
The differences between the brute-force and sophisticated versions are indeed striking from multiple viewpoints. Besides exponential speedup (from n × n to n × log n), there is a proportional increase in complexity. Nonetheless, by focusing on API-centric development, clients of our classes are totally shielded from inner details. Both versions can be used in exactly the same fashion by invoking public methods specified in the base class. This is once again a testimony that sound software engineering skills are real saviors in data science endeavors.
The moral of the story is that purely technological hocus-pocus cannot get you far away. Sure, we demonstrated Numba in the previous chapter, but real breakthroughs can only happen by heralding mathematics. This is generally true for computer science, not only data science, although in case of the latter, the effect is even more amplified, as no naive approach can cope with Big Data.
Besides time-related performance optimization, you may want to measure memory consumption and other resources. For this you also have lots of offerings. Take a look at the memory-profiler package (see https://pypi.org/project/memory-profiler ). There are also suitable magic command wrappers %memit and %mprun. You can use them by first issuing %load_ext memory_profiler. The same is true for time profiling using the line_profiler module (see https://pypi.org/project/line_profiler ) with the %lprun magic command.
Interactive Information Radiators
Most often we produce active reports that summarize our findings regarding some static dataset. A common platform to deliver live reports of this kind is the standard Jupyter notebook. However, in some situations the supporting dataset is continually changing over time. Forcing users to rerun notebooks and pick up the latest dataset isn’t a scalable solution (let alone resending them a refreshed notebook each time). A better option is to set up a pipeline and let dynamic reports be created automatically. This is the idea behind information radiators, which are also known as dashboards.
We can differentiate two major categories of fluctuating data: batch-oriented data, which is accumulated and processed at regular intervals (for example, hourly, daily, weekly, etc.), and streaming data, which is updated continuously. The concomitant dashboard must reflect this rate of change and expose solely crucial business metrics for monitoring and fast decision making. Any superfluous detail will just deter users’ attention. Of course, which type of dashboard you should build also depends on your pipeline’s capabilities.
Current energy production and consumption
Energy production by different plants (nuclear, thermal, water, wind, solar, etc.)
The short-term weather forecast, which is an outside factor that could considerably impact the efficacy of wind and solar plants
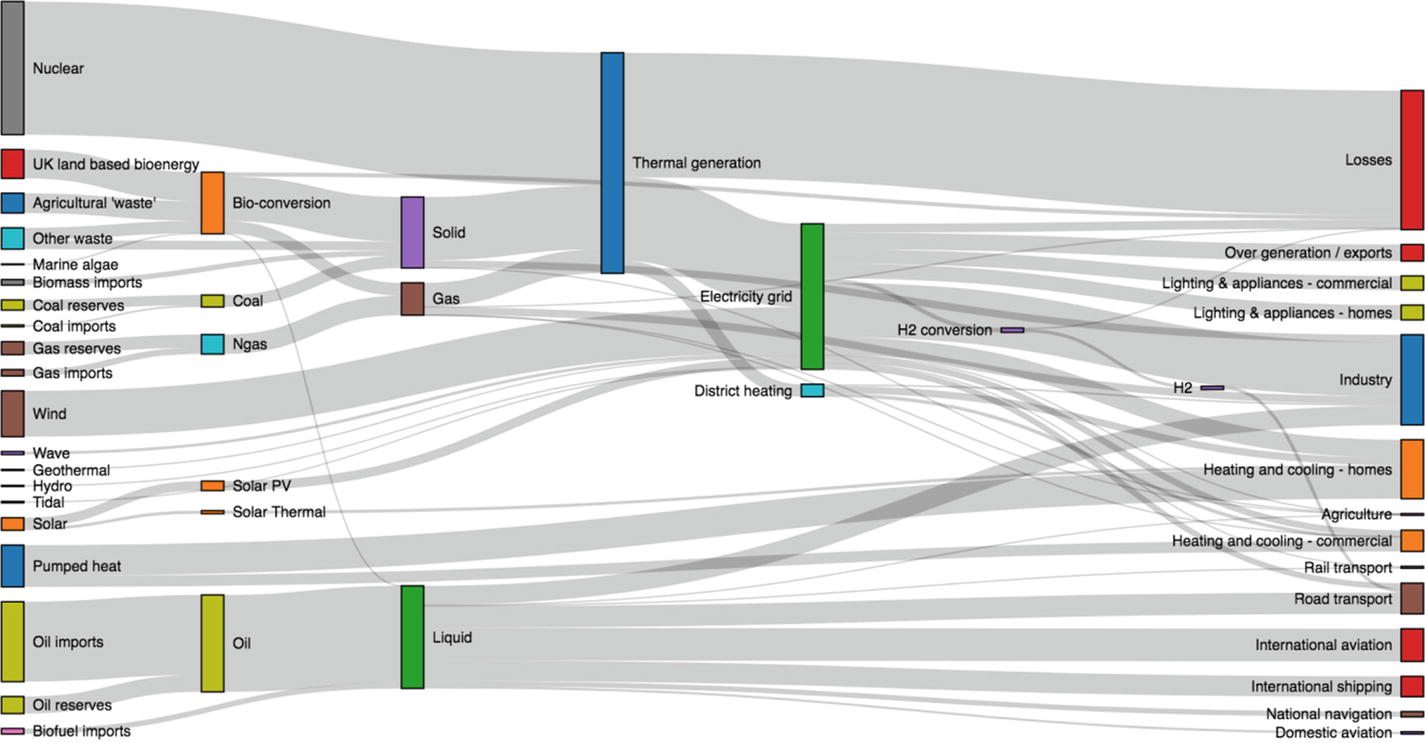
A contrived scenario of energy production and consumption in the UK in the future (see https://github.com/d3/d3-sankey )
The Sankey diagram is by default interactive, and you can select more details about particular branches by clicking or hovering over it. Now imagine this diagram is part of your widely accessible notebook that is regularly updated by your pipeline. This is the core idea behind dashboarding with notebooks (see reference [5]).
Regularly updated and published dataset(s) (for trying out things, you may use some datasets offered by Kaggle; see reference [5])
A dashboarding pipeline that would pick up relevant datasets and rerun your notebook
An interactive notebook, most likely utilizing Altair (see https://altair-viz.github.io ), Plotly (see https://plot.ly ), Bokeh (see https://bokeh.pydata.org ), or a similar framework, as you probably don’t want to code stuff from scratch
The Power of Domain-Specific Languages
Without Any High-level Framework, There Are Lots of Moving Targets to Keep Under Control
Nonetheless, the differences are obvious once you visualize these datasets, as shown in Figure 6-12. Listing 6-9 shows the snippet that achieves a similar visualization, but this time using Altair’s DSL (see also reference [6] about DSLs). The code is compact without unnecessary low-level details. With a DSL, you only specify what you want to accomplish and let the engine decide the best tactic to deliver the requested output. You need to run this code from a JupyterLab notebook by simply pasting it into a code cell.
Visualization Using Altair’s DSL
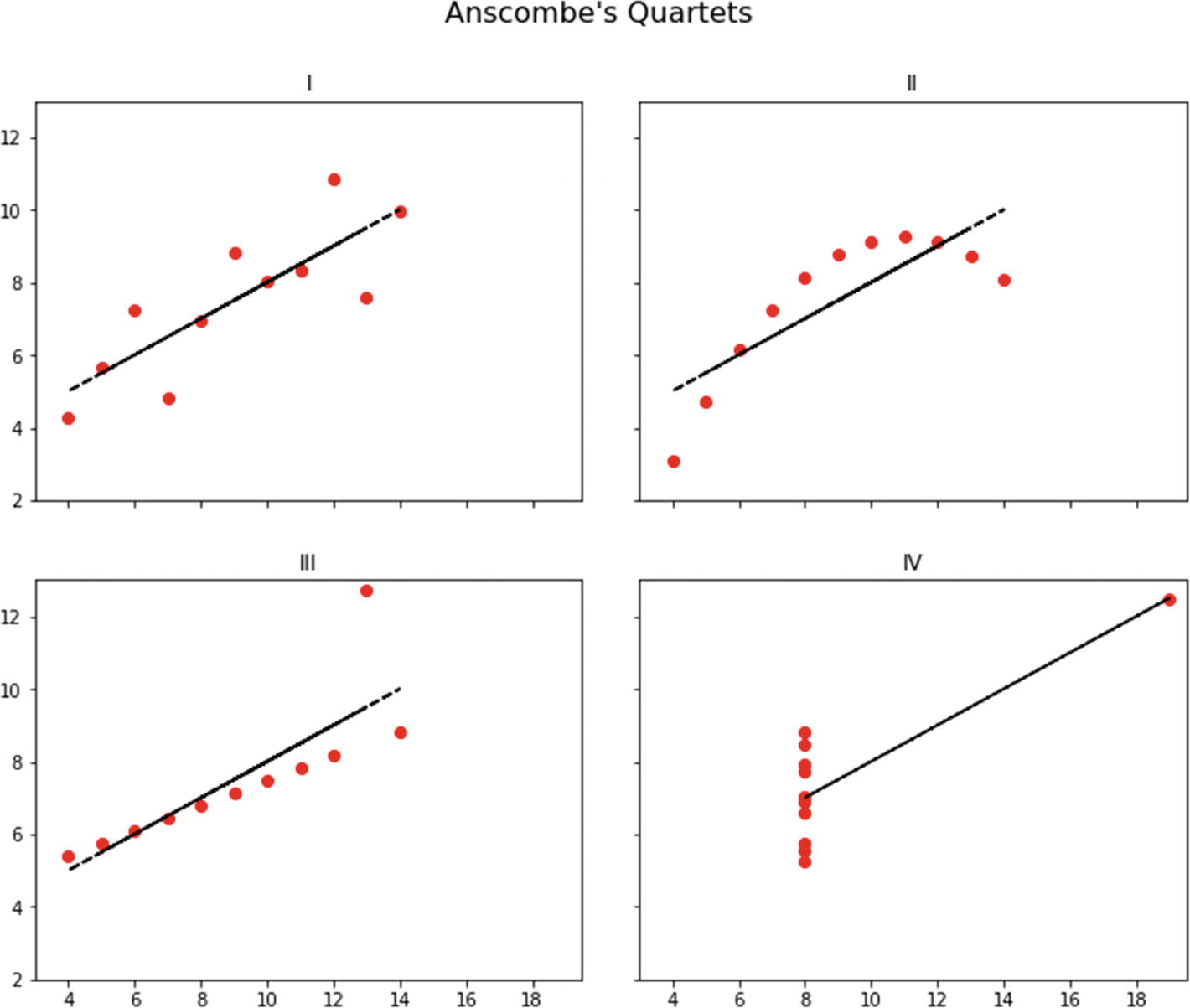
The four quartets are definitely different, although they share the same summary statistics
Tip
You may want to create a snapshot of the dashboard’s state at some moment. All charts produced by Altair include a drop-down menu to save diagrams as images or export them into Vega format.
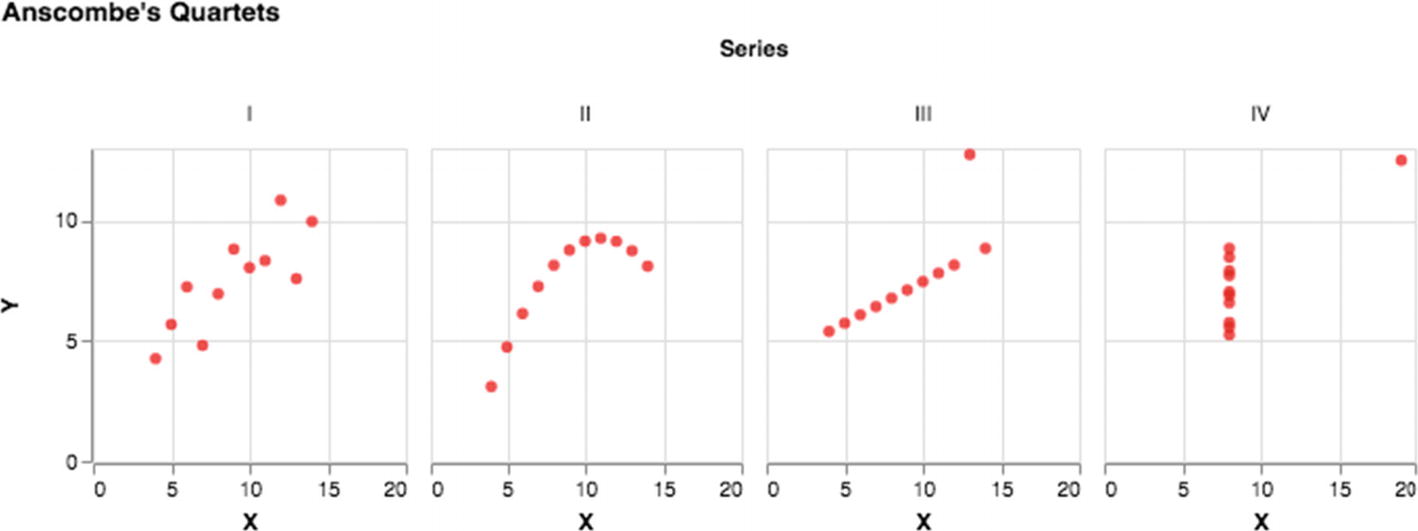
Anscombe’s quartet using default arrangement in Altair
Another advantage of using declarative frameworks is that your notebook will be devoid of device-specific stuff. The whole point of having dashboards is to allow people to access data anytime and everywhere. This entails that many people will use portable devices to browse data. Your dashboard must fit the screen perfectly regardless of whether the user is viewing it on a computer, laptop, tablet, or smart phone. Taking care of all this should be left to the framework.
Exercise 6-1. Create a Factory
Clients shouldn’t instantiate classes directly; ideally, they shouldn’t even be aware of concrete implementation classes. This is the doctrine behind the principle to program against interfaces instead of implementation artifacts. There are two design patterns associated with realizing this vision: Factory Method and Abstract Factory. The latter makes another step in this direction to isolate clients even from factory implementations.
Refactor the code base and introduce these factory entities. In this manner, clients would simply pass the relevant portfolio to a factory (describing the properties of the desired implementation) and receive an object exposed through the shared API (interface).
Exercise 6-2. Implement a Test Harness
- 1.
Get a fresh instance of points by calling the BaseClosestPair.generate_points method (the number of points should be capped at 1000).
- 2.
Find the correct solution using some reliable oracle (for example, by using the trivial NaiveClosestPair class).
- 3.
Call the target implementation with the same dataset.
- 4.
Compare and report the result.
Thanks to the existence of the common API, your test harness may accept any target implementation of this type. This will create an opportunity for future optimization attempts. Try to produce a ParallelClosestPair class. Observe that our algorithm is inherently parallelizable, since left and right groups may be processed independently.
Exercise 6-3. Produce an Ultra-Fast Variant
Thus far, we have used only pure Python constructs. Unfortunately, this does not usually result in a top-performing solution. Lists are very slow, since they are flexible and heterogeneous. Using the built-in array would be a better option for our case. Nonetheless, even this is not enough. Real speedup would happen by switching to Numpy arrays and compiled code.
Create an UltraFastClosestPair class that utilizes Numpy arrays; I recommend that you do Exercise 6-2 before embarking on this exercise. Notice that some stuff will automatically disappear from the current source. For example, Numpy provides the argsort method out of the box. Furthermore, you can use libraries around Numpy. The sortednp package (see https://pypi.org/project/sortednp ) is very handy, as it implements the merge method in O(n) time.
Exercise 6-4. Practice Overlaying Charts in Altair
In our matplotlib-based code, notice how the regression line is combined with the base scatter plot; see the bold plot instruction in Listing 6-4. Everything was bundled together without having the notion of a chart as a separate abstraction. This is completely different in Altair, where charts behave like self-contained entities. If you have handles to multiple charts, then you can mix them in an ad hoc manner utilizing the overloaded + operator.
Add regression lines to the chart referenced from variable base. You would still use Numpy to compute the coefficients. Take a look at the following tutorial for more details: https://altair-viz.github.io/gallery/poly_fit.html . Furthermore, visit https://stackoverflow.com/questions/50164001/multiple-column-row-facet-wrap-in-altair for help on how to simulate subplots from matplotlib.
Summary
It is impossible to iterate over all kinds of visualizations, since there are myriads of them; you may want to visit the D3 visualization gallery at https://github.com/d3/d3/wiki/Gallery , which collects many types of visualizations in a single place. The most important thing to remember is to always take into account the goals of stakeholders and contemplate how graphical presentation of information may help them. In other words, everything you do must have a good reason and be part of the context. Don’t make fancy displays just for the sake of entertainment.
Interactive displays are very powerful, as they combine analytics with graphical presentation. When the underlying data is changing rapidly, users typically like to monitor those changes and perhaps perform actions pertaining to what-if scenarios. Therefore, center your interaction around those frequent activities, so that users may quickly play around with data in a focused manner. Making powerful interactive dashboards demands knowledge and proficiency about user experience design, which is quite extensive territory. Again, we have revisited the same conclusion that any nontrivial data science project is a team effort.
Dashboarding requires automation, and cloud-based services are well suited for this purpose. They provide the necessary scalable and fault-tolerant infrastructure so that you can set up your pipeline smoothly. You might want to look into two prominent options: PythonAnywhere (see https://www.pythonanywhere.com ) and Google Cloud Platform (see https://cloud.google.com ). The former relies on Amazon Web Services (AWS). Of course, you can also create an on-premises solution using OpenStack (see https://www.openstack.org ). You can quickly get a jump-start on dashboarding by using Kaggle’s API and guidelines from reference [5].
References
- 1.
Michael Dubakov, “Visual Encoding,” https://www.targetprocess.com/articles/visual-encoding , Sept. 2012.
- 2.
Sanjoy Dasgupta, Christos Papadimitriou, and Umesh Vazirani, Algorithms, McGraw-Hill Education, 2006.
- 3.
Thomas H. Cormen, Charles E. Leiserson, Ronald L. Rivest, and Clifford Stein, Introduction to Algorithms, Third Edition, MIT Press, 2009.
- 4.
Tom Cormen and Devin Balkcom, “Algorithms,” Khan Academy, https://www.khanacademy.org/computing/computer-science/algorithms .
- 5.
Rachael Tatman, “Dashboarding with Notebooks: Day 1,” https://www.kaggle.com/rtatman/dashboarding-with-notebooks-day-1/notebook , 2019.
- 6.
Martin Fowler, Domain-Specific Languages, Addison-Wesley Professional, 2010.
