The Shark-ML library also implements the OCSVM algorithm for anomaly detection. In this case, the OneClassSvmTrainer and the KernelExpansion classes implement the algorithm. The following example shows how it works:
UnlabeledData<RealVector> data;
importCSV(data, dataset_name);
// separate last two samples in test dataset
data.splitBatch(0, 50);
auto test_data = data.splice(1);
double gamma = 0.5; // kernel bandwidth parameter
GaussianRbfKernel<> kernel(gamma);
KernelExpansion<RealVector> ke(&kernel);
double nu = 0.5; // parameter of the method for controlling the
//smoothness of the solution
OneClassSvmTrainer<RealVector> trainer(&kernel, nu);
trainer.stoppingCondition().minAccuracy = 1e-6;
trainer.train(ke, data);
double dist_threshold = -0.2;
RealVector output;
auto detect = [&](const UnlabeledData<RealVector>& data) {
for (size_t i = 0; i < data.numberOfElements(); ++i) {
ke.eval(data.element(i), output);
if (output[0] > dist_threshold) {
// Do something with anomalies
} else {
// Do something with normal
}
}
};
detect(data);
detect(test_data);
First, we loaded the object of the UnlabeledData class from the CSV file and split it into two parts: one for training and one for testing. Then, we declared the kernel object of the GaussianRbfKernel type and initialized an object of the KernelExpansion class with it. The KernelExpansion class implements an affine linear kernel expansion. This can be represented with the following formula:
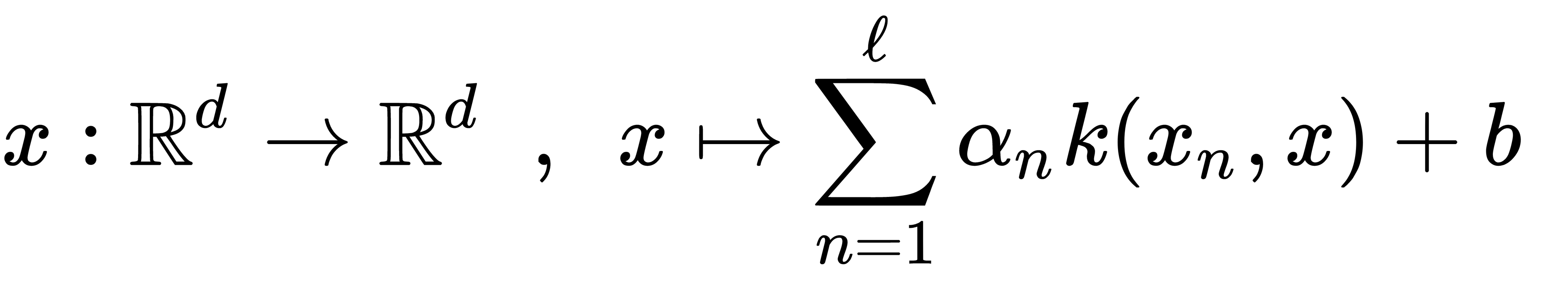
Using this object's type is a requirement defined by the Shark-ML API, but we can use it for a more precise configuration of the algorithm. After we put the kernel expansion object in place, we initialized an object of the OneClassSvmTrainer class and configured it. We also configured the stopping criteria and the solution smoothness parameter. Then, we used the train() method to fit this algorithm to our training data. After training was completed, we used the eval() method of the KernelExpansion object to detect anomalies. This method returns values that we can interpret as distances from the class boundary. By doing this, we can compare them with the threshold.
