In the previous chapter, I explored the idea that most data scientists often come from a background that is quite far removed from traditional computer science/software engineering. Consequently, they produce code that is perfectly suitable for great exploratory data analysis, statistical modeling, or innovative ML experiments but not robust enough for the production phase of a large business platform. Data scientists often think in terms of the next analysis script but not along the lines of the next software module that integrates into a larger system.
Scripting is (mostly) the code you write for yourself. Software is the assemblage of code you (and other teammates) write for others. It is an undeniable fact that most data scientists, not having a traditional software development background and training, tend to write AI/ML analysis code mostly for themselves.
They just want to get to the heart of the pattern hidden in the data. Fast. Without thinking deeply about normal mortals (users). They write a block of code to produce a rich and beautiful plot. But they don’t create a function out of it to use later. They import lots of methods and classes from standard libraries. But they don’t create a subclass of their own by inheritance and add methods to it for extending the functionality.
In the previous chapter, you explored some of these issues through scikit-learn code and a typical classical ML task, fitting a logistic regression model. In this chapter, you will explore how similar principles can help you write better code for deep learning tasks with some hands-on examples using Keras/TensorFlow.
Modular Code and Object-Oriented Style for Productive DL
Functions, inheritance, methods, classes: they are at the heart of robust object-oriented programming (OOP). But you may not want to delve deeply into them if all you want to do is create a Jupyter notebook with your exploratory data analysis and plots.
You can avoid the initial pain of using OOP principles, but this almost always renders your notebook code non-reusable and non-extensible. More precisely, that piece of code serves only you (until you forget what exact logic you coded) and no one else. But readability (and, thereby, reusability) is critically important for any good software product/service. That is the true test of the merit of what you produce. Not for yourself. But for others.
Data science involving deep learning models and code is no exception. These days, powerful and flexible frameworks like TensorFlow or PyTorch make the actual coding of a complex neural network architecture relatively simple and brief. However, if the overall DS code is not modularized and well-organized (following much of the style discussed in Chapter 4 in the section “Why (and How) to Modularize Code for Machine Learning”), then it is plagued by the same issues of non-reproducibility and non-reusability. Let’s see some examples of how you can organize and modularize DL code in your data science work.
Example of a Productive DL Task Flow
One of the most common and repetitive tasks for DL analysis is to build out a deep neural network (DNN) object. Data scientists routinely use non-modularized code to just add layers (e.g., from Keras (https://keras.io/api/layers/) or PyTorch https://pytorch.org/tutorials/recipes/recipes/defining_a_neural_network.html) APIs) and build this as a local variable in their Jupyter notebook. Wouldn’t it be a much better idea to create a custom function for this task?
After building an (untrained) model, you must compile (set learning rate, batch size, etc.) and run the model with data. Would a custom function help make this task modularized as well?
When you make such a DNN builder function, which parameters will be passed on? Which ones can be optional? What are the default values? If you encounter a situation where you don’t know how many parameters need to be passed on, are you using the *args and **kwargs that Python offers?
Did you write a docstring for that function to let others know what the function does and what parameters it expects plus an example?
Can you also modularize the code used to create the visual analytics based on the output of those model functions?

Deep learning task flow organized in modular fashion
Wrappers, Builders, Callbacks
Fundamentally, in the subsection above I described wrapping up the most essential tasks in a DL-based workflow inside custom functions and using them as the core building blocks of your data science code. Additionally, you can wrap up the tasks related to data formatting/transformation and prediction/inference in a similar fashion.
It is to be noted that wrapper functions for regression and classification tasks can have separate sets of architecture and parameters. So, it makes sense to keep their build customized. The choice of the default parameter values in the wrapper functions is of critical importance, too.
Early stopping based on some error or computation criterion
Periodically saving the model to disk (making the system robust against unexpected failure)
Obtaining an overview on various internal states and statistics of a model in mid-flight (i.e., while the training is going on)
Finally, if you want to extend this approach all the way to the full OOP paradigm, you can build out classes and utility modules incorporating all these wrappers as special methods. You can call this a DL utility module, which you can call upon in any data science task where supervised ML modeling is needed.
Modular Code for Fast Experimentation

A slice of the Fashion MNIST dataset
Business/Data Science Question
The basic ML task for this dataset seems straightforward. But what if there is a higher-order optimization or visual analytics question around this core ML task: how does the model architecture complexity impact the minimum epochs it takes to reach the desired accuracy?
It should be clear to you why we even bother about such a question: because this is related to the overall business optimization. Training a neural net is not a trivial computational matter (www.technologyreview.com/s/613630/training-a-single-ai-model-can-emit-as-much-carbon-as-five-cars-in-their-lifetimes/). Therefore, it makes sense to investigate what minimum training effort must be spent to achieve a target performance metric and how the choice of architecture impacts it.
The image classification accuracy could be related to a broader business outcome such as a fashion recommendation or clothing identification in a store. The core data science task helps optimize the cost of running that business task—to use the image database with the optimal expenditure of computing resources using the ML code as the underlying nuts and bolts.
In this example, you will not even use a convolutional neural network (CNN; https://towardsdatascience.com/a-comprehensive-guide-to-convolutional-neural-networks-the-eli5-way-3bd2b1164a53), which are commonly used for image classification tasks. This is because, for this dataset, a simple densely connected neural net can accomplish reasonably high accuracy, and, in fact, a sub-optimal performance is required to illustrate the main point of the higher-order optimization question posed above.
What the minimum number of epochs for reaching the desired accuracy target and how do you determine this?
How does the specific architecture of the model impact this number or training behavior?
Creating an inherited class from a base class object
Creating utility functions and calling them from a compact code block that can be presented to an external user for higher-order optimization and analytics
Inherit from the Keras Callback

A custom class built on top of a Keras callback

Snapshot of an example run with the callback enabled
Model Builder and Compile/Train Functions

Snapshot of a model builder function
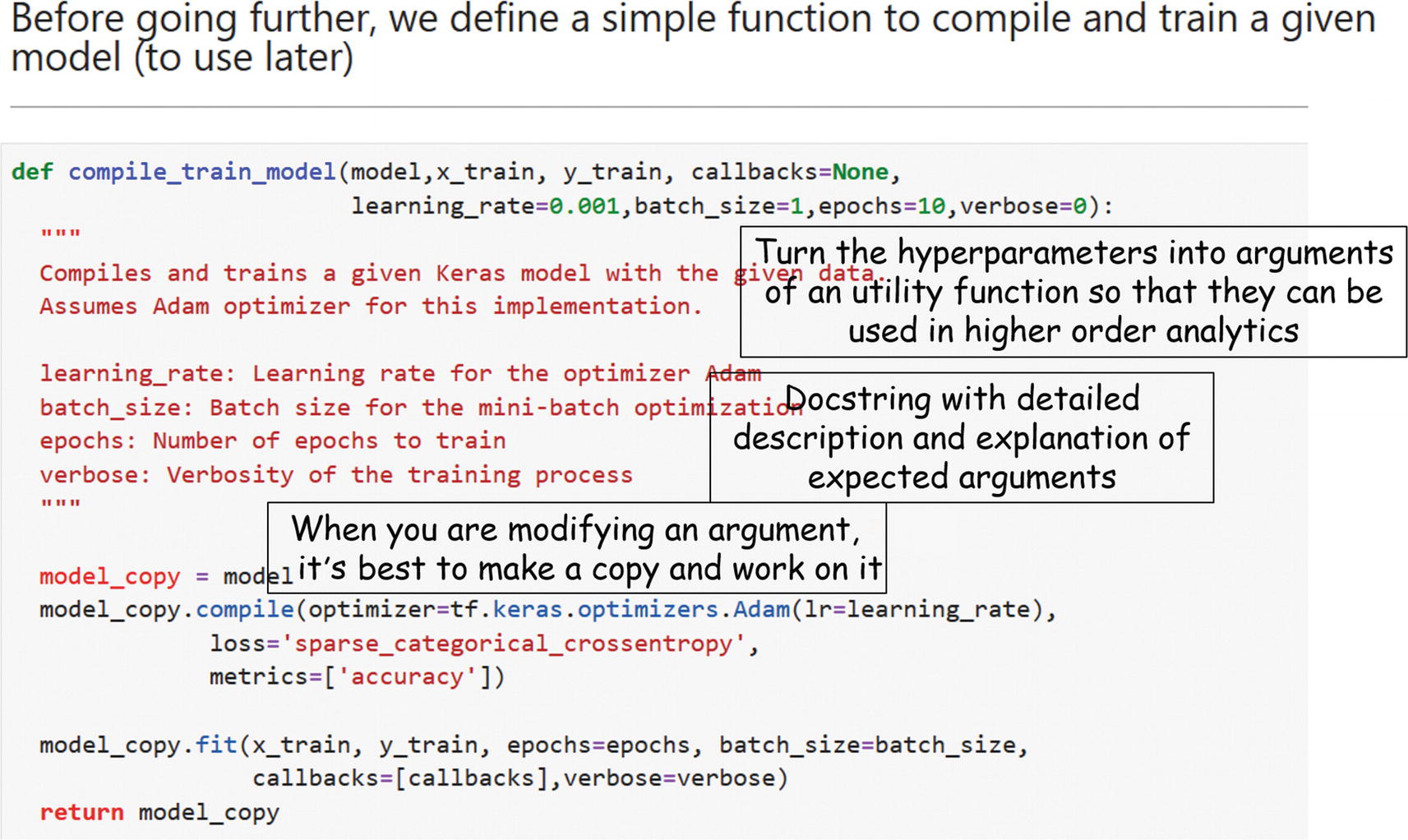
Snapshot of a model compiling and training function
Visualization Function

Snapshot of the visualization function

A typical loss-accuracy plot from the trained DL model
Final Analytics Code, Compact and Simple
Thus far you have modularized the core DL code. Now you can take advantage of all the functions and classes you defined earlier and bring them together to accomplish the higher-order optimization task. Consequently, your final code will be highly compact, but it will generate the same interesting plots of loss and accuracy over epochs for a variety of accuracy threshold values and DNN architectures (neuron counts).
This will give you the ability to use a minimal amount of code to produce visual analytics about the choice of performance metric (classification accuracy in this case) and DNN architecture. This is the first step towards building an optimized machine learning system.

Some representative cases are generated for the optimization task
The final analytics/optimization code is succinct and easy to follow for a high-level user who does not need to know the complexity of Keras model building or callbacks classes. This is the core principle behind OOP, the abstraction of the layers of complexity, which you are able to accomplish for your deep learning task.

Representative results for various model architecture (neuron counts per hidden layer) and accuracy targets
Also, note the custom titles for each plot. These titles clearly enunciate the target performance and the complexity of the neural net, thereby making the analytics easy. It was a small addition to the plotting utility function, but this shows the need for careful planning while creating such functions. If you had not planned for such an argument to the function, it would not have been possible to generate a custom title for each plot. This careful planning of the API (application program interface) is part and parcel of good OOP.
Turn the Scripts into a Utility Module

Building a deep learning utility module (for your own use)
Summary of Good Practices
You just learned some good practices, borrowed from OOP, to apply to a DL analysis task. Almost all of them may seem trivial to seasoned software developers. However, this chapter is designed for budding data scientists who may not have that structured programming background but need to understand the importance of imbuing these good practices in their ML workflow.
Whenever you get a chance, turn repetitive code blocks into utility functions.
Think very carefully about the API of the function (i.e., the minimal set of arguments required and how they will serve a purpose for a higher-level programming task).
Don’t forget to write a docstring for a function, even if it is a one-liner description.
If you start accumulating many utility functions related to the same object, consider turning that object to a class and the utility functions as methods.
Extend class functionality whenever you get a chance to accomplish complex analysis using inheritance.
Don’t stop at Jupyter notebooks. Turn them into executable scripts and put them in a small module. Build the habit of modularizing your work so that it can be easily reused and extended by anyone, anywhere.
In the next chapter, you will try your hand at building your own ML estimator class based on these principles. For a taste of DL utility functions and a neural net trainer class, please read “Deep learning with Python” at https://github.com/tirthajyoti/Deep-learning-with-Python/tree/master/utils.
Streamline Image Classification Task Flow
Image classification is one of the most common tasks in a data science workflow involving deep learning tools. Streamlining or automating such a task is, therefore, a prime example of the automation and modularization that I have been preaching thus far.
For this specific task, a data scientist may desire a single function to automatically pull images from a specified directory on the disk (or from a network address) and give back a fully trained neural net model, ready to be used for prediction. Therefore, in this section, you will explore how to use a couple of utility methods from the Keras (TensorFlow) API to streamline the training of such models (specifically for a classification task) with built-in data preprocessing.
Grab some data.
Put it inside a directory/folder arranged by classes.
Train a neural net model with minimum code/fuss.

Streamlining (and simplifying) the image classification task
The Dataset
Daisy
Tulip
Rose
Sunflower
Dandelion
For each class, there are about 800 photos. The photos are not particularly high resolution (about 320 x 240 pixels each). They are not reduced to a single size since they have different proportions. However, they come organized neatly in five directories named with the corresponding class labels. You can take advantage of this organization and apply the Keras methods to streamline the training of your convolutional network.
The full Jupyter notebook is in the GitHub repository. I will use selected snapshots of the code in this section to show the important parts for illustration.
It is recommended to run this script on a GPU. You will build a convolutional neural net (CNN) with five convolutional layers; consequently, the training process with thousands of images can be computationally intensive and slow if you are not using some sort of GPU. For the Flowers dataset, a single epoch took ~1 minute on my laptop with a NVidia GTX 1060 Ti GPU (6GB Video RAM), Core i-7 8770 CPU, and 16GB DDR4 RAM.
Building the Data Generator Object
This is where the actual magic happens. The official description of the ImageDataGenerator class says "Generate batches of tensor image data with real-time data augmentation. The data will be looped over (in batches)."
Basically, it can be used to augment image data with a lot of built-in preprocessing such as scaling, shifting, rotation, noise, whitening, etc. Right now, you’ll just use the rescale attribute to scale the image tensor values between 0 and 1. Here is a useful article on this aspect of the class: “How to increase your small image dataset using Keras ImageDataGenerator”(https://medium.com/@arindambaidya168/https-medium-com-arindambaidya168-using-keras-imagedatagenerator-b94a87cdefad).
But the real utility of this class for the current demonstration is the super useful method named flow_from_directory, which can pull image files one after another from the specified directory. Note that this directory must be the top-level directory where all the subdirectories of individual classes can be stored separately. The flow_from_directory method automatically scans through the subdirectories and sources the images along with their appropriate labels.
You can specify the class names (as you did here with the classes argument) but this is optional. However, you will later see how this can be useful for selective training from a large trove of data.
Another useful argument is the target_size, which lets you resize the source images to a uniform size of 200 x 200, no matter the original size of the image. This is some cool image-processing right there with a simple function argument.
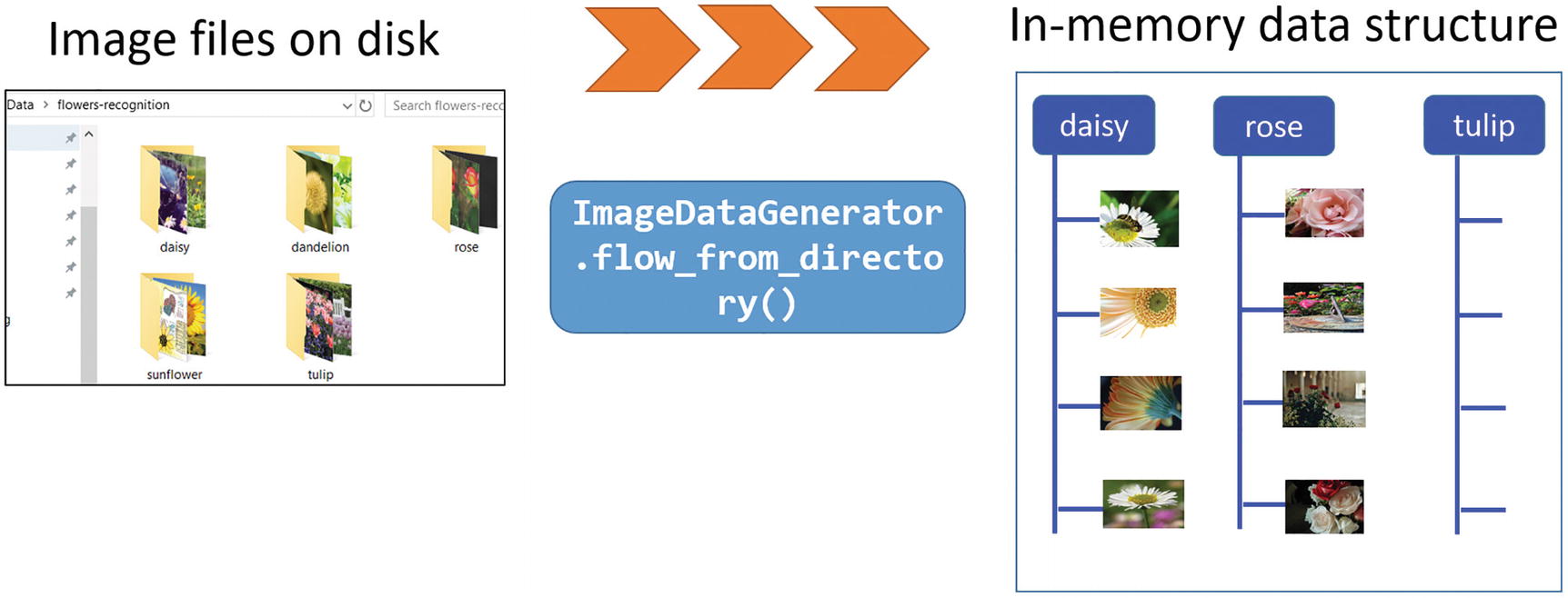
The ImageDataGenerator object working on the Flower dataset
What’s more interesting is that this is also a Python generator object (https://realpython.com/introduction-to-python-generators/). That means it will be used to yield data one by one during the training. This significantly reduces the problem of dealing with a very large dataset whose contents cannot be fitted into memory at one go.
Building the Convolutional Neural Net Model

Summary of the CNN model used for flower classification
Training with the fit_generator Method
I discussed the cool things the train_generator object does with the flow_from_directory method and with its arguments. Let’s utilize this object in the fit_generator method of the CNN model, defined above.

Representative loss/accuracy plots of the CNN training task
Encapsulate All of This in a Single Function
What have you accomplished so far?
You have been able to utilize the Keras ImageDataGenerator and fit_generator methods to pull images automatically from a single directory, label them, resize and scale them, and flow them one by one (in batches) for training a neural network.
Can you encapsulate all of this in a single function?

Encapsulate the core components in a single function. The user supplies a directory name and gets back a trained model
When you are designing a high-level API, you should aim for more generalization than what is required for a particular demo. With that in mind, you can think of providing additional arguments to this function to make it applicable to other image classification cases (you will see an example soon).
train_directory: The directory where the training images are stored in separate folders. These folders should be named as per the classes.
target_size: Target size for the training images. A tuple such as (200,200).
classes: A Python list with the classes for which you want the training to happen. This forces the generator to choose specific files from the train_directory and not look at all the data.
batch_size: Batch size for training
num_epochs: Number of epochs for training
num_classes: Number of output classes to consider
verbose: Verbosity level of the training, passed to the fit_generator method

Snapshot of the single utility function that streamlines the classification task
Testing the Utility Function
You test the train_CNN function by simply supplying a folder/directory name and getting back a trained model that can be used for predictions. Suppose that you want to train only for daisy, rose, and tulip classes and ignore the other two flowers’ data. You simply pass on a list to the classes argument. In this case, you must set the num_classes argument to 3.
You will notice how the steps per epoch are automatically reduced to 20 as the number of training samples is less than the case above. Also, note that verbose is set to 0 by default in the function above, so you need to specify explicitly verbose=1 if you want to monitor the progress of the training epoch-wise.
Does It Work (Readily) for Another Dataset?
This is an acid test for the utility of such a function: can we just take it and apply to another dataset without much modification? Let’s find out.

The Caltech-101 image dataset
The Caltech-101 dataset was built by none other than famous Stanford professor Dr. Fei Fei Li (https://profiles.stanford.edu/fei-fei-li) and her colleagues (Marco Andreetto and Marc Aurelio Ranzato) at Caltech in 2003 when she was a graduate student there. We can surmise, therefore, that Caltech-101 was a direct precursor for her work on ImageNet.

Directory of the stored Caltech-101 images
All you did is to pass on the address of this directory to the function and choose the categories of the images you want to train the model for. Let’s say you want to train the model for classification between cup and crab. You can just pass their names as a list to the classes argument as before.
Also, note that you may have to reduce the batch_size significantly for this dataset as the total number of training images will be much lower compared to the Flowers dataset, and if the batch_size is higher than the total sample, you will have steps_per_epoch equal to 0 and that will create an error during training.

Training happening with Caltech-101 images (two classes, cup and crab)
The model predicted the class correctly for the crab test image.
You can download any random image and test the performance of your model. If not satisfied, you should train the model by changing the architecture and hyperparameters using the modularized function.
The main point, however, is that you were able to train a CNN model with just the same two lines of code for a completely different dataset than you started with. This is the power of modularizing code and building a generic API that works with a wide variety of data sources. This saves valuable time and makes the code reusable. The edifice of productive data science stands on these foundational elements.
Other Extensions
So far, inside the fit_generator you only had a train_generator object for training. But what about a validation set? It follows exactly the same concept as a train_generator. You can randomly split from your training images a validation set and set it aside in a separate directory (the same subdirectory structures as the training directory) and you should be able to pass that on to the fit_generator function.
Want to directly work with a pandas DataFrame that stores your image? No problem. There is a method called flow_from_dataframe for the ImageDataGenerator class where you can pass on the names of the image files as contained in a pandas DataFrame and the training can proceed.
You are strongly encouraged to check out and extend these ideas as you see fit for your applications.
Activation Maps in a Few Lines of Code

The black-box problem of deep learning (source: CMU ML blog, https://blog.ml.cmu.edu/2019/05/17/explaining-a-black-box-using-deep-variational-information-bottleneck-approach/).
This does not bode well because we humans are visual creatures (www.seyens.com/humans-are-visual-creatures/). Millions of years of evolution have gifted us an amazingly complex pair of eyes (www.relativelyinteresting.com/irreducible-complexity-intelligent-design-evolution-and-the-eye/) and an even more complex visual cortex (www.neuroscientificallychallenged.com/blog/know-your-brain-primary-visual-cortex), and we use these organs to make sense of the world. The scientific process starts with observation, and that is almost always synonymous with vision. In business, only what we can observe and measure can we control and manage effectively. Seeing/observing is how we start to make mental models (https://medium.com/personal-growth/mental-models-898f70438075) of worldly phenomena, classify objects around us, separate a friend from a foe, and so on.
Activations maps have been proposed to help visualize the inner workings of complex CNN models. Let’s talk about them.
Activation Maps
Several approaches for understanding and visualizing CNNs have been developed in the literature, partly as a response to the common criticism that the learned internal features in a CNN are not interpretable. The most straightforward visualization technique is to show the activations of the network during the forward pass.
At a simple level, activation functions help decide whether a neuron should be activated. This helps determine whether the information that the neuron is receiving is relevant for the input. The activation function is a non-linear transformation that happens over an input signal, and the transformed output is sent to the next neuron.
Activation maps are just a visual representation of these activation numbers at various layers of the network as a given image progresses through as a result of various linear algebraic operations. One can deduce the workings of the network and design limitations from these maps. For ReLU activation-based networks, the activations usually start out looking relatively blobby and dense, but as the training progresses the activations usually become sparser and more localized. One design pitfall that can be easily caught with this visualization is that some activation maps may be all zero for many different inputs, which can indicate dead filters and can be a symptom of high learning rates.
However, visualizing these activation maps is a non-trivial task, even after you have trained your neural net well and are making predictions out of it. How do you easily visualize and show these activation maps for a reasonably complicated CNN with just a few lines of code?
Activation Maps with a Few Lines of Code
In the previous section, I showed how to write a single compact function to obtain a fully trained CNN model by reading image files one by one automatically from the disk. Now you’ll you use this function along with a nice little library called Keract, which makes the visualization of activation maps very easy. It is a high-level accessory library of Keras to show useful heatmaps and activation maps on various layers of a neural network.
Therefore, for this code, you need to use a couple of utility functions from the module you built earlier, train_CNN_keras and preprocess_image, to make a random RGB image compatible for generating the activation maps.
You’ll use the same Caltech-101 dataset discussed in the last section. However, you are training only with five categories of images: crab, cup, brain, camera, and chair.
Training
To generate the activations, you can choose a random image of a human brain from the Internet or any other source. Store the test image as the file brain-1.jpg.
Activation

Activation map arrays are stored (the variable length corresponding to the size of the convolutional filter at that layer)

Activation maps for the first convolution layer

Activation maps for the second convolution layer
For this model, there are 5 convolutional layers (followed by max pooling layers), so you get back 10 sets of images. For brevity, I won’t show the rest, but you are encouraged to explore and see them by playing with the Jupyter notebook.
Another Library for Web-Based UI
Another beautiful library for activation visualization is called Quiver. However, this one is built on the Python microserver framework Flask and displays the activation maps on a browser port rather than inside your Jupyter Notebook. It also needs a fully trained Keras model as input. So, you can easily use the utility function described in the previous section and try this library for interactive visualization of activation maps.
How Is This Productive Data Science?
In this chapter, you learned how by using only a few lines of code (utilizing compact functions from a special module and a nice little accessory library to Keras) you can train a CNN, generate activation maps, and display them layer by layer—from scratch. This gives you the ability to train CNN models (simple to complex) from any image dataset (as long as you can arrange them in a simple directory format) and look inside their guts for any test image you want.
And once you build the necessary utility modules and the activation map scripts, you can reuse and apply them to a wide variety of image data. This leads to a fast and efficient exploration of a large set of images for all kinds of applications. This is why this kind of approach integrates with the story of productive and efficient data science.
Hyperparameter Search with Scikit-learn
Keras is one of the most popular go-to Python libraries/APIs for beginners and professionals in deep learning. Although it started as a stand-alone project by François Chollet, it has been integrated natively into TensorFlow starting in version 2.0. Read more about it here (https://keras.io/about/):. As per its own official doc, it is “an API designed for human beings, not machines” as it “follows best practices for reducing cognitive load.”
Now, hyperparameter tuning is one of the situations where the cognitive load is sure to increase. DL models have a great many hyperparameters to begin with: learning rate, decay rate, activation function, dropout rate, momentum, batch size, and more. Optimizing a DL model for best performance and computing cost depends critically on the right choice of these hyperparameters. Therefore, data scientists spend a lot of time and effort tuning them manually or via some automated script or optimization strategy/framework.
Although there are many supporting libraries and frameworks for handling it, for simple grid searches, Keras offers a beautiful API to integrate with our favorite scikit-learn library. In this section, we will talk about it.
Scikit-learn Enmeshes with Keras
Almost every Python machine-learning practitioner is intimately familiar with the scikit-learn library and its beautiful API with simple methods (www.tutorialspoint.com/scikit_learn/scikit_learn_estimator_api.htm) like fit, get_params, and predict. The library also offers extremely useful methods for cross-validation, model selection, pipelining, and grid search abilities. Data scientists use these tools for classical ML problems every day. But can you use the same APIs for a deep learning problem?
It turns out that Keras offer a couple of special wrapper classes, both for regression and classification problems, to utilize the full power of these APIs that are native to scikit-learn. In this section, you will work using a simple k-fold cross-validation (https://medium.com/datadriveninvestor/k-fold-cross-validation-6b8518070833) and exhaustive grid search with a Keras classifier (www.tensorflow.org/api_docs/python/tf/keras/wrappers/scikit_learn/KerasClassifier) model. It utilizes an implementation of the scikit-learn classifier API for Keras.
Data and (Preliminary) Keras Model
You tackle a simple binary classification task using the popular Pima Indians Diabetes dataset (www.kaggle.com/uciml/pima-indians-diabetes-database). This dataset is originally from the National Institute of Diabetes and Digestive and Kidney Diseases (www.niddk.nih.gov/). The objective of the dataset is to diagnostically predict whether or not a patient has diabetes, based on certain diagnostic measurements included in the dataset.
You do some minimal data preprocessing including scaling the feature data with MinMaxScaler from scikit-learn. You can pass this X_scaled vector to the special wrapper class you will create.
The KerasClassifier Class
Note how you pass on your model creation function as the build_fn argument. This is an example of using a function as a first-class object in Python (https://dbader.org/blog/python-first-class-functions) where you can pass on functions as regular parameters to other classes or functions.
For now, you have fixed the batch size and the number of epochs you want to run your model for because you just want to run cross-validation on this model. Later, you will treat them as hyperparameters and do a full grid search over them to find the best combination.
Cross-Validation with the Scikit-learn API
The output variable cv_results is a NumPy array consisting of all of the accuracy scores. Accuracy is the metric you coded in your model compiling process. Obviously, you could have chosen any other classification metric like precision or recall, and in that case, that metric would have been calculated and stored in the cv_results array.
You can easily calculate the average and standard deviation of the 10-fold CV run to estimate the stability of the model predictions. This is one of the primary utilities of a cross-validation run and now you can gauge the stability of any Keras model using this approach.
Grid Search with a Updated Model
Activation function
Optimizer type
Initialization method
Batch size
Number of epochs
Make the exhaustive hyperparameter search space size as 3 × 3 × 3 × 3 × 3 = 243. Note that the actual number of Keras runs will also depend on the number of cross-validation you choose, as cross-validation will be used for each of these combinations. In total, there will be 729 fittings of the model, 3 cross-validation runs for each of the 243 parametric combinations. If you don’t like the full grid search, you can always try a randomized grid search.

Exhaustive grid search options
You set the cv = 3 to reduce the time for the run. By default, it will be set to 5 by scikit-learn if you leave out that argument.
It is advisable to set the verbosity of GridSearchCV to 2 to keep visual track of what’s going on. Remember to keep verbose=0 for the main KerasClassifier class, though, as you probably don't want to display all the gory details of training individual epochs.

Fitted grid search estimator with all the parameters

Snapshot of best hyperparameter choice printed
You did the initial 10-fold cross-validation using ReLU activation and Adam optimizer and got an average accuracy of 0.691. After doing an exhaustive grid search, you discover that a tanh activation and a rmsprop optimizer could have been better choices for this problem.

DataFrame created from the grid search parameters

Visualizations of the grid search results
Summary
This chapter covered a variety of topics centered on the idea of making commonly used deep learning code and tasks more productive and efficient. I carried over the idea of modularizing the code from the previous chapter and showed hands-on examples with useful model building and plot functions with the Keras framework. A powerful construct called the Keras callback was also discussed in this context.
Next, I discussed the idea of streamlining one of the most common DL tasks that a data scientist can encounter: image classification. The goal was to arrive at a single utility function that presents a very simple API to the user. You just pass on a folder name to this function, and it will return a fully trained conv net model by processing all the images in that folder. Not only did you build this function step by step, but you also demonstrated the utility of such an API by applying it to a completely different dataset.
In the next section, you further utilized this function and integrated it with a special library that can extract and visualize activation maps for the various convolution layers of the DL model. Basically, you demonstrated how to visualize the inner workings of a complex DL model with only a few lines of code. Together, these two sections embodied the true journey towards productive and efficient data science involving deep learning.
Finally, you explored the topic of making hyperparameter search easy and seamless. Although there are many dedicated libraries and frameworks for this task, you saw a simple and intuitive approach using the grid search tool from scikit-learn and some special wrapper classes from Keras. It also demonstrated how two of the most popular ML libraries, Keras and scikit-learn, can work together in a seamless manner.
Making deep learning code and products fast and efficient is a huge topic by itself. There are countless approaches and research directions focusing on this. This chapter only aims to induce some fundamental ideas so that you can explore them further.


