6
Handling Unbalanced Data in Clinical Images
Amit Verma*
School of Computer Science, UPES, Dehradun, Uttarakhand, India
Abstract
Manual detection of abnormalities accurately in the clinical images like MRIs by the operators is tedious work that requires good experience and knowledge, specifically manually segmenting the brain tumor in the MRI for further diagnosis by the doctor. So, multiple automatic and semi-automatic approaches were developed to automate the process of segmenting the malignant area of the tumor. The major problem which arises to train the model for automatic segmentation of clinical images is the imbalanced data set. An imbalanced clinical data set means the healthy tissues are always far greater than the cancerous tissues. This difference between the majority and minority data in the data sets reduces or adversely affects the accuracy of predicting model due to biased training data sets. So, it becomes a major concern for the various researchers to balance the data before using it to train a particular prediction model, and various data-level and algorithm–levelbased approaches were developed to balance the imbalance data for improving the accuracy of the trained model. In this chapter, the concept and problem of imbalanced data are discussed and various approaches for balancing the data are also highlighted in which one of the state-of-the-art method bagging is discussed in detail.
Keywords: Bagging, unbalanced data, boosting, MRI, deep learning, medical, brain tumor
6.1 Introduction
Magnetic Resonance Images (MRIs) play a vital role in detecting brain tumors; in India, it is a fast-growing disorder and mainly in children [1]. In India, Glioblastoma Multiforme (GBM), Meningioma, Astrocytoma, etc., are some of the most common categories of a malignant brain tumor [2–6]. MRIs are clinical images that can be considered pre-diagnosis for any patient with brain abnormality in size, functioning, detecting tumor, or anatomy. MR imaging allows operators to take brain pictures on various radio frequencies for better analysis of the area of interest [7]. But it always remains a challenging task to manually segment the brain tumor due to its uneven or irregular size and shape [33, 34]. Operators manually segment the tumor size using some graphical tools to make the report, and further, the doctor manually analyzes the MRI based on the report of the operator for diagnosis. Semi-automatic and automatic segmentation of MRI remains a matter of concern for various researchers, and many state-of-the-art methods [8–15] have been developed to automate the procedure of segmenting the brain tumor on MR images with higher accuracy so that doctor can diagnose the patient in a much better way. The most common problem for applying machine learning or deep learning to train the model for automating the process of segmenting MR images is imbalanced data.
Imbalanced data means when the data is biased to one or more specific class [16, 17], let say an MRI of the brain with malignant tumor will have far more pixels representing the healthy tissues than the cancerous one. Therefore, the model, directly train based on MRI images with such data, will get biased toward the healthy tissues and can predict cancerous tissues as healthy. The imbalanced data set trained model drastically decreases the performance of the predictor, and the class imbalance is one of the common problems in segmenting the clinical images (MRIs). For solving the problem of imbalanced data set, many algorithm-level and data-level–based methods were introduced. The approach of data-level–based methods is to re-sampling and under-sampling of a positive and negative class of data [18, 19]. Whereas in algorithm-level–based methods [20–22], modifications were done in learning algorithm to handle imbalanced data in learning data set to train the predictor. Cluster-based under-sampling [23] is one of the state-of-the-art methods under data-level–based methods for handling imbalanced data. Bagging (Bootstrap aggregating) [24] is one of the most popular approaches for handling the problem of imbalanced data, which is an algorithm-based technique.
In this chapter, the problem of imbalanced data and the consequences of training the model based on imbalanced data are explained in simple language. Further, two main approaches that are cluster-based and bagging considering the clinical data are discussed. The objective of this chapter is to make the reader understand the problem of imbalanced data in segmenting the MR images mainly for brain tumor detection and to get the knowledge about the two state-of-the-art methods for solving the problem of making a biased model.
6.2 Handling Imbalance Data
The problem of class imbalance is one of the major concerns in segmenting the clinical images majorly like segmenting the brain tumor. As the data sets containing the spatial resolution information based on the MRIs of the brain with tumor mostly have large data (pixels) representing the healthy tissues of the brain as compare to the malignant area. In simple words, Figures 6.1 and 6.2 represent the distribution of imbalanced and balanced data over a 2D graph [25]. As we can see that blue dots which are in majority in Figure 6.1 can be considered as pixels representing the healthy tissues and red dots representing the cancerous ones, and blue dots are far more than red which is representing the biased data for creating the predictor model. So, the trained model based on imbalanced data will have low accuracy and can misclassify the cancerous tissue as health which could be more costly for the patient than the wrong classification of healthy tissue as cancerous [26]. So, it is important to balance the data before using it for training the model. The example of balanced data is shown in Figure 6.2 where blue dots representing the healthy tissues are almost the same as the malignant tissues represented by red dots. A variety of state-of-the-art methods have been developed to improve the efficiency of the predictor model by correcting the problem of imbalanced data. Majorly, the overall work by various researchers for balancing the data set can be classified into two main categories that are performing over-sampling or under-sampling on the data that is called at data-level–based technique [18], and modifying the learning algorithm according to the biased data is called the algorithm-level–based technique [19]. Some of the notified approaches under data-level–based techniques are random cluster–based under-sampling [27], random over-sampling–based method [28], SMOTE [29], and MSMOTE [30], and major algorithm-level–based techniques are bootstrap aggregation method [24], boosting approach [31], and adaptive booting [32], which are some of the notable techniques for handling the imbalance data. Cluster-based under-sampling and bagging are discussed in detail in the following subsections.

Figure 6.1 Typical example of imbalanced data.
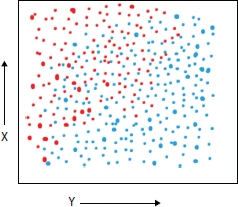
Figure 6.2 Typical example of balanced data.
6.2.1 Cluster-Based Under-Sampling Technique
Classification of a class is the process of predicting the class for unknown data with the help of trained model; it is well-known technique in machine learning [37–39]. Classifier is trained with the training data set and the hypotheses model is prepared to get the result for some new inputs. There are vast application of classification analysis. Any classification model broadly involve four major steps that are collection of sample data, distribute the data in training data and testing data keeping in mind that training data instances should always be greater than the testing data, train the model using training data, and now model is ready to predict the result for some new data. But the major concern is always the accuracy of the predictor model which majorly depends on the uniformity of the training samples. If the training data is not uniform, this means uniform distribution of labeled data than the model (predictor) could be get biased toward majority data. This arises the problem of imbalance class distribution.
For balancing the data, under-sampling approach is adopted by many researchers for its better accuracy as compare to the over-sampling approach. Considering the data of brain tumor MR images for segmenting the malignant part in the image. Assuming that HT is the majority data of healthy tissue in the data set and MT, the data with minority data represent the malignant tissues. Now to maintain the uniformity in the data, HT is randomly divided into several subsets of data as shown below.
The number of distributed subsets depends on the variance between the size of HT and MT; let the size of HT be H and the size of MT be L. Then, the number of subsets would be S according to the equation below.
where
With each subset, L will be concatenated.
For example, if H = 1,000 and L = 10, then the recommended number of subsets S would be 100 and with each set Si, L would be concatenated for final set Sf .
Now, the data is balanced and can be used to train the predictor using any modeling technique for segmenting the brain tumor in the MR images.
For more detailed specification, let the total size of the data D be 1,100 in which HT = 1,000 and MT = 100. Now, we make four clusters of the data set D as shown in Table 6.1, showing the data in majority HT and minority MT data sets.
Table 6.1 Cluster distribution.
| Cluster ID | HT | MT | Si = HTi/MTi |
| 1 | 300 | 30 | 10 |
| 2 | 300 | 30 | 10 |
| 3 | 200 | 20 | 20 |
| 4 | 200 | 20 | 10 |
Now, according to the equation below, the majority samples are selected randomly and concatenated with the minority sample for final data sets to train the model [40].
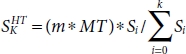
where m is the ratio of HT:MT and k is the total number of clusters. So, the final number of majority data sample to be selected in each cluster and concatenated with the minority samples are shown in Table 6.2.
Now, each cluster with the majority sample is concatenated with minority samples MT.
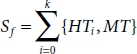
Table 6.2 Numbers of majority sample in each cluster.
| Cluster ID | HTi |
| 1 | 1 * 100 * 10 / (10 + 10 + 20 + 20) = 17 |
| 2 | 1 * 100 * 10 / (10 + 10 + 20 + 20) = 17 |
| 3 | 1 * 100 * 10 / (10 + 10 + 20 + 20) = 34 |
| 4 | 1 * 100 * 10 / (10 + 10 + 20 + 20) = 34 |
With the above equation, we get the final balanced data set to train the model for the prediction.
6.2.2 Bootstrap Aggregation (Bagging)
Bagging [24] is one of the notable algorithm-level–based approaches for handling unbalanced data, majorly used for clinical data such as MRI data of brain tumor segmentation. For classifying the tissues with higher accuracy, considering the BRATS2015 data set [35] tissues can be segmented as either healthy or cancerous with four different categories. The four different categories (labeled) of malignant tissues are edema, non-enhancing (solid), non-enhancing (core), and necrotic [36]. Bagging technique can be used with imbalanced data D for better classification accuracy. In the process of bagging, multiple data sets {d1, d2, ...., dm} are dragged from the given unbalanced data D as shown in Figure 6.3, and in each data set, data is randomly drawn from the data D with replacement. By the term with replacement, we mean that already drawn elements from D can be drawn again. It is recommended that training instances in data sets should be 60% of training instances in D. Now, on various data sets from d1 to dm, training models either same or different are applied on their data sets. Each model gives an output from y1 to ym, and an average of all outputs are taken for better accuracy or the process of voting is adopted to get the class of data, which is given as output by a maximum number of models.
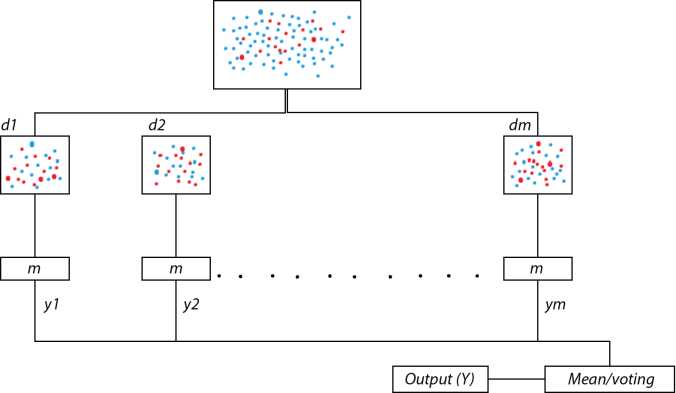
Figure 6.3 The process of bootstrap aggregation.
Bagging is more effective with unstable models like classification and linear regression as in unstable procedures small changes in D can result in a larger change in the predictor model, whereas bagging degrades the performance with the stable procedure like k-nearest neighbor method.
6.3 Conclusion
In this chapter, we discuss the importance of balancing the imbalanced data and how the model trained using imbalance data can affect the performance of the predictor. Imbalanced data is the common and major problem with clinical data like the MRI data for brain tumor segmentation. Considering the problem of imbalanced data, data with the high variance between the majority and minority data. In the case of MRI data for brain tumor segmentation, data representing the healthy tissues are mostly always much higher than the data representing the malignant tissues. So, for making a predictor model to automatic segmentation, the balancing of data is required. The process of bagging is discussed in the chapter for handling imbalanced data.
References
1. Hndu, Over 2,500 indian kids suffer from brain tumour every year, The Hindu, New Delhi, India, 2018, www.thehindu.com/sci-tech/health/Over-2500-Indian-kids-suffer-from-brain-tumour-every-year/article14418512.ece (2018). LastAccessed: June 2019.
2. Drevelegas, A. and Nasel, C., Imaging of brain tumors with histological correlations, Springer Science & Business Media, New York, 2010.
3. Dupont, C., Betrouni, N., Reyns, N., Vermandel, M., On image segmentation methods applied to glioblastoma: state of art and new trends. IRBM, 37, 3, 131–143, 2016.
4. Bauer, S., Wiest, R., Nolte, L.P., Reyes, M., A survey of mri-based medical image analysis for brain tumor studies. Phys. Med. Biol., 58, 13, R97, 2013.
5. Menze, B.H., Jakab, A., Bauer, S., Kalpathy-Cramer, J., Farahani, K., Kirby, J., Burren, Y., Porz, N., Slotboom, J., Wiest, R. et al., The multimodal brain tumor image segmentation benchmark (brats). IEEE Trans. Med. Imaging, 34, 10, 1993–2024, 2015.
6. Mohan, G. and Subashini, M.M., Mri based medical image analysis: survey on brain tumor grade classification. Biomed. Signal Process. Control, 39, 139–161, 2018.
7. Drevelegas, A. and Nasel, C., Imaging of brain tumors with histological correlations, Springer Science & Business Media, New York, 2010.
8. Agn, M., Puonti, O., Law, I., af Rosenschöld, P., van Leemput, K., Brain tumor segmentation by a generative model with a prior on tumor shape, in: Proceeding of the multimodal brain tumor image segmentation challenge, pp. 1–4, 2015.
9. Corso, J.J., Sharon, E., Dube, S., El-Saden, S., Sinha, U., Yuille, A., Efficient multilevel brain tumor segmentation with integrated bayesian model classification. IEEE Trans. Med. Imaging, 27, 5, 629–640, 2008.
10. Menze, B.H., Van Leemput, K., Lashkari, D., Weber, M.A., Ayache, N., Golland, P., A generative model for brain tumor segmentation in multimodal images, in: International conference on medical image computing and computer-assisted intervention, Springer, pp. 151–159, 2010.
11. Prastawa, M., Bullitt, E., Ho, S., Gerig, G., A brain tumor segmentation framework based on outlier detection. Med. Image Anal., 8, 3, 275–283, 2004.
12. Bauer, S., Nolte, L.P., Reyes, M., Fully automatic segmentation of brain tumor images using support vector machine classification in combination with hierarchical conditional random field regularization, in: International conference on medical image computing and computer-assisted intervention, Springer, pp. 354–361, 2011.
13. Hamamci, A., Kucuk, N., Karaman, K., Engin, K., Unal, G., Tumor-cut: segmentation of brain tumors on contrast enhanced mr images for radiosurgery applications. IEEE Trans. Med. Imaging, 31, 3, 790–804, 2012.
14. Lun, T. and Hsu, W., Brain tumor segmentation using deep convolutional neural network, in: Proceedings of BRATS-MICCAI, 2016.
15. Pratondo, A., Chui, C.K., Ong, S.H., Integrating machine learning with region-based active contour models in medical image segmentation. J. Vis. Commun. Image Represent., 43, 1–9, 2017.
16. He, H. and Garcia, E.A., Learning from imbalanced data. IEEE Trans. Knowl. Data Eng., 21, 9, 1263–1284, 2009.
17. López, V., Fernández, A., García, S., Palade, V., Herrera, F., An insight into classification with imbalanced data: Empirical results and current trends on using data intrinsic characteristics. Inf. Sci., 250, 113–141, 2013.
18. Shelton, C.R., Balancing multiple sources of reward in reinforcement learning, in: Advances in Neural Information Processing Systems, T.K. Leen, T.G. Dietterich, V. Tresp (Eds.), vol. 13, pp. 1082–1088, MIT Press, Cambridge, MA, USA, 2001.
19. Jang, J., Eo, T., Kim, M. et al., Medical image matching using variable randomized undersampling probability pattern in data acquisition, in: International Conference on Electronics, Information and Communications, pp. 1–2, 2014.
20. Sudre, C.H., Li, W., Vercauteren, T. et al., Generalised dice overlap as a deep learning loss function for highly unbalanced segmentations, in: Deep Learning in Medical Image Analysis and Multimodal Learning for Clinical Decision Support, New York, pp. 240–248, Springer, 2017.
21. Rezaei, M., Yang, H., Meinel, C., Deep neural network with l2-norm unit for brain lesions detection, in: International Conference on Neural Information Processing, Springer, pp. 798–807, 2017.
22. Hashemi, S.R., Salehi, S.S.M., Erdogmus, D. et al., Tversky as a loss function for highly unbalanced image segmentation using 3d fully convolutional deep networks. CoRR, 1–8, abs/1803.11078, 2018.
23. Yen, S.J. and Lee, Y.S., Cluster-based under-sampling approaches for imbalanced data distributions. Expert Syst. Appl., 36, 3, 5718–5727, 2009.
24. Breiman, L., Bagging predictors. Mach. Learn., 24, 2, 123–140, 1996.
25. Small, H. and Ventura, J., Handling unbalanced data in deep image segmentation, University of Colorado, Boulder, CO 80309, United States, 2017.
26. López, V., Fernández, A., García, S., Palade, V., Herrera, F., An insight into classification with imbalanced data: Empirical results and current trends on using data intrinsic characteristics. Inf. Sci., 250, 113–141, 2013.
27. Yen, S.J. and Lee, Y.S., Cluster-based under-sampling approaches for imbalanced data distributions. Expert Syst. Appl., 36, 3, 5718–5727, 2009.
28. Moreo, A., Esuli, A., Sebastiani, F., Distributional random oversampling for imbalanced text classification, in: Proceedings of the 39th International ACM SIGIR conference on Research and Development in Information Retrieval, pp. 805–808, 2016, July.
29. Chawla, N.V., Bowyer, K.W., Hall, L.O., Kegelmeyer, W.P., SMOTE: synthetic minority over-sampling technique. J. Artif. Intell. Res., 16, 321–357, 2002.
30. Hu, S., Liang, Y., Ma, L., He, Y., MSMOTE: Improving classification performance when training data is imbalanced, in: 2009 second international workshop on computer science and engineering, 2009, October, vol. 2, IEEE, pp. 13–17.
31. Guo, H. and Viktor, H.L., Learning from imbalanced data sets with boosting and data generation: the databoost-im approach. ACM Sigkdd Explor. Newsl., 6, 1, 30–39, 2004.
32. Taherkhani, A., Cosma, G., McGinnity, T.M., AdaBoost-CNN: An adaptive boosting algorithm for convolutional neural networks to classify multi-class imbalanced datasets using transfer learning. Neurocomputing, 404, 351–366, 2020.
33. Shivhare, S.N., Kumar, N., Singh, N., A hybrid of active contour model and convex hull for automated brain tumor segmentation in multimodal MRI. Multimedia Tools Appl., 78, 24, 34207–34229, 2019.
34. Shivhare, S.N. and Kumar, N., Brain tumor detection using manifold ranking in flair mri, in: Proceedings of ICETIT 2019, Springer, Cham, pp. 292–305, 2020.
35. Kistler, M., Bonaretti, S., Pfahrer, M., Niklaus, R., Büchler, P., The virtual skeleton database: an open access repository for biomedical research and collaboration. J. Med. Internet Res., 15, 11, e245, 2013.
36. Havaei, M., Davy, A., Warde-Farley, D., Biard, A., Courville, A., Bengio, Y., Larochelle, H., Brain tumor segmentation with deep neural networks. Med. Image Anal., 35, 18–31, 2017.
37. del-Hoyo, R., Buldain, D., Marco, A., Supervised classification with associative SOM, in: International Work-Conference on Artificial Neural Networks, pp. 334–341, Springer, Berlin, Heidelberg, 2003, June.
38. Lee, T.S. and Chen, I.F., A two-stage hybrid credit scoring model using artificial neural networks and multivariate adaptive regression splines. Expert Syst. Appl., 28, 4, 743–752, 2005.
39. Li, X., Ying, W., Tuo, J., Li, B., Liu, W., Applications of classification trees to consumer credit scoring methods in commercial banks, in: 2004 IEEE International Conference on Systems, Man and Cybernetics (IEEE Cat. No. 04CH37583), vol. 5, pp. 4112–4117, IEEE, 2004, October.
40. Yen, S.J. and Lee, Y.S., Under-sampling approaches for improving prediction of the minority class in an imbalanced dataset, in: Intelligent Control and Automation, pp. 731–740, Springer, Berlin, Heidelberg, 2006.
- *Email: [email protected]
