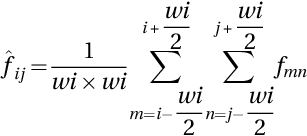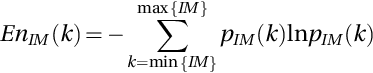Advanced neutrosophic set-based ultrasound image analysis
Deepika Koundal; Bhisham Sharma Department of Computer Science and Engineering, Chitkara University School of Engineering and Technology, Chitkara University, Himachal Pradesh, India
Abstract
Ultrasound (US) images have recently become more widespread because of their better quality, noninvasive and nonionizing radiation, and portability for medical applications. Ultrasound is mostly preferred for initial medical monitoring, diagnosis, and follow-up in comparison to other medical image modalities for its low cost. Because of its benefits, it is broadly utilized for the diagnosis of various diseases in the liver, breast, prostate, and thyroid as well as carotid atherosclerosis. In spite of this, US suffers from poor quality and many artifacts. This makes the extraction of useful information from US images a challenging task for diagnosis, which makes it an active area of research. The issues are being overwhelmed by neutrosophic based methods that are presented for detection, classification, and characterization of abnormalities in different types of organs. This chapter discusses some of the advances in neutrosophic sets (NSs) using US images covering various organs. The focus is on the methodologies, algorithms, and computer-aided diagnosis systems developed by several researchers. By this, various techniques-based on NSs are discussed with their pros and cons for US images for disease diagnosis in clinical practice.
Keywords
Ultrasound; Neutrosophy; Images processing; Segmentation; Denoising; Classification
1 Introduction
Diagnostic Ultrasound (US) has become popular for its high quality, economy, safe nature, and portability. This modality is very attractive due to its noninvasive and nonionizing radiation for medical applications. It is very economical in comparison to other modalities of medical imaging, which makes it broadly accepted for follow-up, diagnosis, and medical monitoring. It is extensively employed for the diagnosis of various types of diseases found in the breast, liver, prostate, thyroid, and coronaries as well as carotid atherosclerosis, in addition to obstetrics and cardiology. However, ultrasound images are affected by a low signal-to-noise ratio, artifacts, and poor quality. An active field of research as well as a challenging task is to extract the valuable information from ultrasound images for diagnosis. Many problems are being overcome and several schemes have been introduced for abnormality detection, characterization, and diagnosis in various types of organs.
For diagnosis of diseases, ultrasound is the most commonly used imaging modality as it is very safe, accessible, portable, user-friendly, and does not use any ionizing radiation (Bosch & Tuinman, 2018; Koundal, Gupta,& Singh, 2012a, 2012b). Ultrasound provides an apparent image of soft tissues such as the thyroid, liver, and kidney that cannot be displayed well on X-ray images (Wells & Liang, 2011). A standard ultrasound system consists of the transducer for scanning, a signal processing device, and a display device. The basic idea is to emanate signals and to gather reflected echoes on the video display screen (Wells, 1999). The transducer is a hand-held gadget about the size of a microphone connected via cord to the scanner. The transducer utilizes a collection of piezoelectric components to transmit high-frequency echoing waves and to receive the sound waves from the scattering structures (McDicken, 1976). When the transducer is compressed against the skin, it produces small inaudible sound waves into the body (Szabo, 2004). As the sound waves reflect from tissues, internal organs, and fluids, the microphone in the transducer accounts for small variations in the direction and pitch of sound waves. When a sound pulse hits an object, it echoes or bounces back. By these echoes, it is possible to find the distance, shape, size, and consistency (solid or filled with fluid) of an object. In the imaging system, the amplitude of each reflected wave is denoted by a dot. The brightness of the dot signifies the power of the returned echo. The location of the dot denotes the depth from which the returning echo was received. These dots are combined to produce a whole image. It gives real-time imaging for directing invasive procedures such as FNA and biopsies. In medicine, ultrasound is used to find variations in the growth of tissues and vessels and to locate abnormal tissues such as nodules as well as their size and contours. The thyroid, breast, liver, kidney, etc., are appropriate for ultrasound study due to their superficial location, vascularity, size, and echogenicity. The majority of nodules are benign and pose no health hazard, so their identification is of great significance to evade needless biopsies. If the nodules are malignant, they will require further diagnosis or prognosis. Thus, the sonographic appearance of a nodule-based on size or the presence of multiple nodules may aid in making decisions for performing biopsies.
The granular appearance, known as a speckle, is a significant feature of the ultrasound image. Speckle noise is an interference pattern that is fully deterministic if the transducer settings and the location of all scatterers in the medium is known (Gupta, Chauhan, & Sexana, 2004). It is also known as multiplicative noise and is difficult to remove as compared to traditional additive Gaussian noise (Huang & Yang, 2013; Lee, Yen, & Ueng, 2012). Artifacts are the result of physical properties of the ultrasound itself, which occur commonly in the ultrasound display. The removal of artifacts is necessary for accurate ultrasound analysis, as artifacts may lead to unnecessary concern or clinical intervention. Artifacts in ultrasound imaging may be categorized into four main types: falsely perceived objects, missing structures, structures with a misregistered location, and degraded images. These artifacts occur from noise or incorrect anatomical imaging. An incorrect display of anatomy during imaging can cause reverberation, speed displacement, and shadowing artifacts (Feldman, Katyal, & Blackwood, 2009; Hindi, Peterson, & Barr, 2013).
Ultrasound images have fuzziness due to speckle noise, vague nodule boundaries, and low contrast between suspicious and surrounding tissues. The nodule boundaries in ultrasound are indistinct and difficult to differentiate because of artifacts. These artifacts cause images of poor quality and low contrast with blurred, indeterminate, and ambiguous edges. Therefore, segmentation of a nodule is a challenging chore due to the low contrast and uncertainty caused by one of the artifacts that is known as speckle noise in ultrasound images. Thus, the main concern is speckle noise. Speckle can be considered as noise as it causes fuzzy, ambiguous, and vague structures in tissues under observation. It is also known as a multiplicative noise that degrades the quality of US images by appearing as arbitrary bright and dark spots.
A number of techniques have been proposed using neutrosophic sets (NSs). NSs are widely used in various types of medical image-processing applications. Several authors have introduced several neutrosophic based techniques for denoising, segmentation, classification, etc.. It has been observed from the literature that numerous denoising methods on the basis of NSs have been given for removing salt and pepper noise as well as Rician, Gaussian, and speckle noise (Guo, Cheng, & Zhang, 2009; Koundal, Gupta,& Singh, 2013, 2016b; Mohan, Chandra,Krishnaveni, & Guo, 2012, 2013; Mohan, Krishnaveni, & Guo, 2011, 2012, 2013b, 2013c; Qi, Liu, & Xu, 2016). For denoising, a neutrosophic based γ-median has been presented for removing Gaussian noise (Mohan, Krishnaveni, & Guo, 2012).
Further, Guo et al. (2009) have presented a denoising scheme-based on NS for removing Gaussian and salt and pepper noise using several variances. Qi et al. (2016) have also introduced a neutrosophic based pixel-wise adaptive method for removing salt and pepper noise. Mohan et al. have given a neutrosophic based wiener filter for removing Rician noise (Mohan, Chandra, et al., 2012, 2013; Mohan, Krishnaveni, et al., 2011, 2012, 2013b; Mohan, Krishnaveni, & Huo, 2015). Furthermore, the neutrosophic based KUAN (NKUAN) method and the neutrosophic LEE (NLEE) method have been presented for speckle noise removal (Koundal et al., 2012a). Another neutrosophic based speckle removal method on the basis of Gamma and Nakagami noise distribution has also been presented (Koundal et al., 2016b). Bajger, Ma, and Bottema (2009) utilized neutrosophic based denoising methods for the denoising of mammograms. For facial recognition problems, another neutrosophic based preprocessing method has been introduced.
In the literature, a number of authors have reported different types of NS-based image segmentation methods. Zhang, Zhang, and Cheng (2010a) reported a neutrosophy-based watershed segmentation approach. Cheng and Guo (2008) presented the NS-based thresholding method for segmenting natural and artificial images. Later, Anter, Hassanien, ElSoud, and Tolba (2014) improved the segmentation algorithm by integrating neutrosophic and fuzzy c-means for CT images. Guo and Sengur (2015a) presented a neutrosophic c-means clustering (NCM) method for data partitioning, particularly indistinct and fuzzy data. Its efficiency was tested on both image segmentation and data clustering applications.
Further, a neutrosophic evidential C-means clustering scheme was introduced with the incorporation of the Dezert–Smarandache theory for the segmentation of images (Guo & Sengur, 2015b). Another NS-based improved fuzzy C-means (IFCM) scheme has been presented for the segmentation of images (Guo & Sengur, 2015b). Akhtar, Agarwal, and Burjwal (2014) introduced a neutrosophic k-means clustering for image segmentation. Karabatak, Sengur, and Guo (2013) have given another neutrosophic based segmentation algorithm for color images with improved parameters and entropy-based criteria. Another method called iterative neutrosophic lung segmentation for lung and rib segmentation has been introduced on the basis of expectation maximization analysis and morphological operations (EMM). Zhang et al. (2010a) presented a region merge method to segment natural images for resolving oversegmentation. Sengur and Guo (2011) have presented another automatic method that combined the color with texture information in the wavelet and neutrosophic domains for segmentation of images (Mathew & Simon, 2014). A neutrosophic similarity clustering (NSC) was presented for segmenting images (Guo, Şengür, & Ye, 2014). A NS-based on an improved artificial bee colony approach was introduced for segmenting synthetic aperture radar (SAR) images (Hanbay & Talu, 2014). A neutrosophy-based unsupervised algorithm for color image segmentation has been reported. An unsupervised segmentation method was introduced that synthesized the neutrosophic set and mean shift (NS-MS) (Yu, Niu, & Wang, 2013).
An automatic segmentation approach was introduced for segmenting jaw lesions in X-ray images. It involved noise removal and a NS-based hybrid fuzzy c-means approach (NFCM) (Alsmadi, 2018). Another segmentation scheme was presented that can correctly and automatically segment the coronary arteries from computed tomography angiography images (Chen et al., 2015). A fully automatic segmentation method based on neutrosophic logic and the watershed method was introduced for the extraction of the liver using CT images, which helped in the diagnosis of liver disease and treatment planning (Sayed, Ali, Gaber, Hassanien, & Snasel, 2015; Siri & Latte, 2017). Furthermore, an unsupervised texture-color image segmentation method with an effective indeterminacy reduction operation has been presented that integrated the nonsubsampled contourlet transform (NSCT) with NS (Heshmati, Gholami, & Rashno, 2016).
A texture-based image segmentation method was introduced with the integration of Gabor filters and a neutrosophic graph cut (NGC). It has been revealed from results that the method has achieved better performance for texture segmentation. The NGC-based image segmentation method was introduced to find qualified rendering images for a thyroid ultrasound. In this, an energy function using neutrosophic values is introduced and segmentation of different anatomic regions is achieved by a maximum flow algorithm (Guo et al., 2017). An indeterminacy filtering and neutrosophic c-means clustering was introduced (Guo, Xia, Şengür, & Polat, 2017). Neutrosophic based segmentation was carried out for tumor detection in an MRI image (Kaur & Kaur, 2016). Further, an image segmentation-based on neutrosophy with the integration of quantum behaved particle swarm optimization (QPSO) was presented (Jianhu, Xiao, Hongmei, Jun, & Xiaomin, 2016; Zhao, Wang, Zhang, Hu, & Jian, 2016). Furthermore, texture features are incorporated with neutrosophic theory for medical image segmentation (Akbulut, Sengür, & Guo, 2016; Koundal, 2017).
Singala and Agrawal (2014) presented a neutrosophic assessment schema for SAR imagery segmentation using swarm optimization techniques. The method provided better results in comparison to swarm optimization algorithms. A neutrosophic cloud detection and localization with wavelet transform was introduced for satellite remote sensing images. This method is efficient for the detection of thin and thick clouds using Landsat images (Mathew, Surya, & Simon, 2013). A computer-assisted diagnosis (CAD) system has been introduced for the classification of breast cancer using thermograms. Further, for the classification of breast parenchyma, different kernel functions were used in a support vector machine (Gaber et al., 2015).
A neutrosophic based image retrieval system has been presented (Eisa, 2014; Rashno & Sadri, 2017). Another neutrosophic segmentation method has been introduced with QPSO (Sayed & Hassanien, 2017). Rashno et al. introduced a fully automatic segmentation method for the segmentation of cysts and fluid-related regions of diabetic macular edema subjects in two-dimensional (2D) optical coherence tomography (OCT). The OCT images are segmented using the NS-based graph shortest path (Rashno et al., 2016; Rashno & Sadri, 2017).
This chapter discusses some of the current neutrosophic based advanced techniques using ultrasound images involving various types of organs. The focus of the chapter is in the techniques, methodologies, and systems introduced by multidisciplinary research teams for CAD.
2 Neutrosophic based CAD system for ultrasound image analysis
2.1 CAD system
Computer-aided diagnosis (CADx) is becoming a popular research area in diagnostic ultrasound imaging. The analysis of ultrasound images suffers from a high interobserver variation rate, as it requires well-trained experienced radiologists and is operator-dependent. Therefore, to reduce the operator-dependent nature and to make the diagnosis practice reproducible, CADx systems are becoming widespread (Chang et al., 2000). There are many advantages of a CADx system, one of which is that it can attain statistical and computational features that cannot be computed intuitively and visually by medical practitioners. Generally, CADx systems for nodule diagnosis in ultrasound images involve various stages such as preprocessing, segmentation, feature extraction, and classification, as shown in Fig. 1.

2.1.1 Preprocessing
Preprocessing of the image is the very first step for improving the quality of ultrasound images. It is used to suppress possible variations that arise during image acquisition or to remove noise or unwanted information from ultrasound images without evading vital information. These variations hinder further image analysis steps.
2.1.2 Image segmentation
Segmentation of images plays a very significant role in the detection of significant regions that are used for the analysis of tissue types, pathological regions, and anatomical structures. The first task is to define a region of interest (ROI) within the organ for eliminating unnecessary regions from processing. After ROI generation, the next task is to segment the disease within the ROI. A correct boundary estimation of a disease helps in further classification and categorization of diseases. Thus, good image segmentation is necessary to maintain the accuracy and sensitivity of the lesion detection and classification system (Guo & Şengür, 2013). After the segmentation of the disease, features can be computed from it to eliminate the false detection rate for accurate and better diagnosis.
2.1.3 Classification
After segmentation, the suspicious areas can be categorized as malignant or benign on the basis of selected features using various classification techniques. The malignant nodules with vague boundaries and distinct histopathological components are often fused with adjoining tissues, making the delineation of tissue a difficult task. To address these issues, the CADx system has crucial importance in segmentation, classification of benign or malignant tissue, and estimation of the volume in ultrasound images (Ju & Cheng, 2013). For this, a fully computerized system is required for improving the accuracy and for decreasing the misdiagnosis rate for earlier detection and diagnosis of diseases.
Presently, researchers are focusing much attention toward neutrosophy-based methods to solve various image-processing problems due to its capability of handling indeterminate information. In the literature, a number of authors have reported different types of NS-based image-processing methods (Bajger et al., 2009; Faraji & Qi, 2013; Salama & Elagamy, 2013). The neutrosophic based denoising and segmentation methods are discussed (Nguyen, Ashour, & Dey, 2019).
2.2 Breast ultrasound image analysis
Shan et al. have presented a segmentation method named the neutrosophic L-means (NLM) clustering method for breast ultrasound (BUS) images (Shan, Cheng, & Wang, 2012). NLM has obtained improved accuracy with fair computational speed. The major constraints of NLM are that it cannot segment more than one lesion and it failed under a severe shadowing effect. Furthermore, a Neutrosophic similarity score (NSS) method is presented with the integration of a level set for breast tumor segmentation on ultrasound images (Guo & Şengür, 2013; Guo, Şengür, & Tian, 2016). The ultrasound image was mapped to the NS domain through membership subsets. Afterward, NSS was utilized to quantify the membership degree of the true region. Eventually, a level set was applied for segmenting the breast tumor in ultrasound images. The experiments showed that NSS can segment the breast tumor accurately and effectively.
Further, a fully automatic, robust, and effective NS-based segmentation method was introduced for BUS images. In this, the ultrasound image is mapped to a binary image, and then the watershed method has been employed for segmenting mapped images to locate the tumor in the segmented area (Zhang, Zhang, & Cheng, 2010b). Further, an approach is presented for the classification of breast nodule characteristics into circumscribed and noncircumscribed classes. The nodule is segmented automatically by integrating the NS and watershed methods along with relevant features extracted from the nodule. The results indicated that the approach had successfully carried out the classification of margin characteristics of the nodule using BUS images. Nugroho, Rahmawaty, Triyani, and Ardiyanto (2017) presented a normalization algorithm using fuzzy c-means with neutrosophic clustering to enhance and segment the image. The method has achieved better performance in segmenting the nodule from BUS images than that without normalization.
Lotfollahi, Gity, Ye, and Far (2018) introduced a neutrosophic based semiautomatic segmentation method. It used the region-based active contour that segmented the BUS images more precisely, even with intensity and inhomogeneity. A nonlocal means filter has been used for removing speckle noise and a fuzzy logic technique has been used for enhancing contrast. This method can be modified for different organs using ultrasound images and is not limited only to BUS images. Classification of lesions into benign and malignant has not been done.
Furthermore, a technique called information gain-based neutrosophic c-means (IGNLM) clustering has been presented (Lal, Kaur, & Gupta, 2018). The technique incorporates the information gain calculated from the local neighborhood for updating the membership values in the NLM clustering process. For clustering decisions about a pixel, the existing NLM method takes into account only its membership value and distance from the cluster center, but pays no attention to the significant characteristics that exist in an image in that the neighboring pixels have similar features and their probability of belongingness to the same cluster is high. This neighborhood information has been exploited in the technique by using a concept of entropy called information gain. It has been subsequently used to improve the segmentation capability of the NLM clustering process. From the results, the technique is fully automatic and robust as it produced homogeneous clustering, even in the presence of shadow regions (Lal et al.). Zhang et al. (2010b) ultrasound image is transformed to the NS domain and then watershed algorithm is employed for segmenting the mapped image. Finally, the tumor is located in the segmented area. Segmentation of the tumor is an essential step for CAD systems of BUS images. As ultrasound images suffer from poor quality, the fuzzy connectedness method failed to segment the objects with weak boundaries. Therefore, neutrosophic connectedness (neutro-connectedness) and neutrosophic subsets have been defined to generalize the fuzzy connectedness and fuzzy subsets. The neutro-connectedness modeled the inherent uncertainty and indeterminacy of the spatial topological features of the image. The method has been evaluated by the average Hausdroff error, the false-positive ratio, and the similarity ratio, as compared to the fuzzy connectedness method. The method is robust and more accurate for the segmentation of tumors in BUS images (Xian, Cheng, & Zhang, 2014).
Experiments have been performed on BUS images to show the applicability of neutrosophic based methods in image denoising and segmentation. One of the visual results on breast cancer using an ultrasound image of the neutrosophic based denoising and segmentation method is shown in Fig. 2. Fig. 2A shows an original image, Fig. 2B illustrates a denoised image, Fig. 2C shows an image of segmented cancer from BUS images, and Fig. 2D illustrates the delineated boundary of cancer using BUS images.

2.3 Thyroid ultrasound image analysis
Furthermore, the NKUAN and the NLEE methods have been presented for speckle noise reduction (Koundal et al., 2012a). The experiments have demonstrated that NS methods performed well as compared to KUAN and LEE on simulated artificial images that are corrupted by speckle with various noise levels. The visual results also revealed that NKUAN and NLEE removed the speckle and preserved the edges. Another method known as the neutrosophic based Nakagami-based total variation (NNTV) method has been presented. The NNTV method transformed the image into the NS domain and employed filtering for removing noise. The method was analyzed qualitatively and quantitatively by evaluation measures and calculating the mean opinion score from three experts on real ultrasound images (Koundal, Gupta, & Singh, 2018). Further, a neutrosophic based nonconvex speckle reduction method based on gamma noise statistics has been introduced to maintain a good balance between texture preservation and speckle suppression (Koundal, Gupta, & Singh, 2016a).
Another automated segmentation method was introduced that combined the spatial neutrosophic clustering with level sets for segmenting thyroid nodules using ultrasound images. The results have shown that it can delineate more than one nodule effectively and accurately (Koundal et al., 2016b). Koundal et al. (2017a) introduced a texture information-based image segmentation method. The cluster center and objective function are updated by integrating texture information in the NS domain. The results verified that the method is able to segment the object more accurately and efficiently. The results are superior to other methods, even in case of images having low contrast and vague boundaries (Koundal, 2017).
The neutrosophic based denoising and segmentation method using a thyroid ultrasound image is illustrated in Fig. 3, where Fig. 3A illustrated the original image, Fig. 3B showed the denoised image, Fig. 3C illustrated the segmented nodule in the thyroid ultrasound image, and Fig. 3D exhibited the marked periphery of the thyroid nodule in the ultrasound image.

2.4 Other ultrasound image analysis
Kaur and Singh (2016) presented the segmentation approach by deploying a NS with fuzzy c-means for a blood vessel. The image is analyzed to check if it is diseased. Diseases are detected with the use of region growing and are classified as cotton exudates, lesions, and wool spots using the neural network classification method. The method is compared with other methods and is tested on the Standard Diabetic Retinopathy Database (DIARETDB1) and the DRIVE database. Koundal et al. (2017b) made a comparison of the NCM clustering and the IFCM clustering, integrating spatial information for segmenting medical images.
3 Ultrasound image in neutrosophic domain
3.1 Image transformation in neutrosophic domain
The three membership functions called true (TM), indeterminate (IM), and false (FM) are the components of a NS that are used for representing 〈A〉, 〈Neut-A〉 and 〈Anti-A〉, respectively. Each pixel in the NS domain has been denoted as PNI = {TM, IM, FM}, where TM, IM, and FM are the likelihoods of pixels that are associated with the set of white, indeterminate, and black pixels, respectively (Guo & Cheng, 2009). The image in the NS domain is illustrated in Fig. 4. The TM membership function can be determined as given below:
where j ranges from 0 to (m − 1, i ranges from 0 to n − 1, ![]() is the minimum gray level value,
is the minimum gray level value, ![]() is the local mean achieved, and
is the local mean achieved, and ![]() is the maximum pixel value.
is the maximum pixel value.
where wi represents the size of the window, fmn signifies the noisy image, and ![]() denotes the local mean of pixels on a window. The membership function IM can be calculated as:
denotes the local mean of pixels on a window. The membership function IM can be calculated as:
where δmin is the minimum absolute difference value, δmax is the maximum absolute difference value, δij is the absolute difference value between pixel values fij and local mean values ![]() . The false membership function FM is determined as
. The false membership function FM is determined as

The true membership TM in the neutrosophic domain is processed by normalization of the gray levels in [0, 1], as represented in Eq. (1). The pixels in ultrasound images may represent the texture information or speckle noise; therefore, it is hard to differentiate. Thus, the neighborhood mean, ![]() , is used for determining the local mean of pixels on a window. The absolute difference, δij, between intensity value,fij, and its local mean value,
, is used for determining the local mean of pixels on a window. The absolute difference, δij, between intensity value,fij, and its local mean value, ![]() , is for computing the indeterminate component of the image. The false membership function, FM, can be calculated as the complement of TM (Guo et al., 2009).
, is for computing the indeterminate component of the image. The false membership function, FM, can be calculated as the complement of TM (Guo et al., 2009).
3.2 Neutrosophic entropy
The entropy in the NS domain, “En”, is employed to measure the indeterminacy degree of images that captured the uncertainities. The En of IM is defined as
The entropy is used to remove the fuzziness and uncertainty presented in images. It evaluated the distribution of gray levels in an image. If the entropy value is less, then the intensities have unequal probability. If the intensities have equal probability, then it is a uniform image. The IM values d measure the indeterminate degree of pixels. The variations in TM influenced the element distribution in IM to make the set IM correlate with TM and vary the entropy of IM.
3.3 Transforming neutrosophic domain to gray level domain
In general, after applying various image-processing operations such as segmentation and denoising, the neutrosophic image is transformed back to the gray level domain by Eq. (7).
where ![]() is the minimum intensity value,
is the minimum intensity value, ![]() is the maximum gray level value, and the
is the maximum gray level value, and the ![]() component is based on IM after the neutrosophic filtering operation.
component is based on IM after the neutrosophic filtering operation.
4 Discussion
Currently, the analysis of ultrasound images has gone through a new era of research aided by advances in neutrosophic theory. The rising commercial and clinical interest is in utilizing portable as well as inexpensive ultrasound devices outside the conventional clinic based settings. This paper highlights some challenges ahead while presenting a perception on this transformation and the probable opportunities in the analysis of ultrasound images that may have high influence on healthcare in the future. Various authors have implemented neutrosophic based techniques on generic images such as the integration of neutrosophic with the Chan–Vese algorithm, principal component analysis, and shearlet transform (Ali, Khan, & Tung, 2018; Amin, Elfattah, Hassanien, & Schaefer, 2014; Ashour, Hawas, Guo, & Wahba, 2018; Cheng, Guo, & Zhang, 2011; Dhar & Kundu, 2017; Guo et al., 2014; Guo, Jiang, et al., 2017; Guo & Şengür, 2014a, 2014b; Guo, Xia, et al., 2017; Hu et al., 2017; Ju & Cheng, 2013; Ponnusamy & Babu, 2016; Qureshi & Ahamad, 2018; Salafian, Kafieh, Rashno, Pourazizi, & Sadri, 2018; Sert & Alkan, 2019; Thanh & Ali, 2017a, 2017b; Zhang, 2010; Zhang & Wang, 2014; Zhang & Zhang, 2015). Most of the work is also carried out on color images using neutrosophy (Guo, Şengür, Akbulut, & Shipley, 2018; Zhang, Zhang, & Cheng, 2012). All this work is required to be implemented using ultrasound images in the future.
Augmented reality- (AR) and virtual reality-based ultrasound will revolutionize medical imaging. AR technologies intermingle the data obtained from different image modalities with real-world aspects. An AR system has been introduced that aids doctors to see real-time data from an ultrasound probe directly into a patient's body through an AR headset rather than on a screen. The system displays from an ultrasound directly. This provides a single unified view instead of splitting the attention of physicians between screen and patient. Physicians generally used a hand-held scanner to determine the blood vessels surrounding the wound. But the AR system helps them in locating those vessels directly by emphasizing them in a 3D image shown in an AR headset.
Another open area in ultrasound image analysis is neutrosophic based image registration and fusion. Image registration and fusion are of great significance in recognizing medical images. A novel method can be drawn to fuse two or more images by using some operations in the neutrosophic domain. The real-time ultrasound image fusion with other imaging modalities will provide priors (statistics on motion or likely shape) that would commence further potential to modify ultrasound images for real-time diagnosis. This will be helpful where large datasets are available and CT/MR images have already attained a component of patient management as it manage the storage capacity, there may be main benefits in taking account of automated fusion of images that are yet to be recognized. Another domain in ultrasound image analysis is Neutrosophic based image classification. In this area, more research is required to be carried out on natural images or disease detection in leafs. Another emerging area is neutrosophic based image retrieval in ultrasound images. This area also requires more work to be done to develop a neutrosophic based retrieval system for ultrasound images. Hybrid methods can be designed by integrating neutrosophic domain methods with other methods such as wavelets to show their effectiveness over other domains. Table 1 lists the advantages and disadvantages of various neutrosophic based methods for different types of tissues using ultrasound images.
Table 1
| References | Methods | Tissue | Advantages | Disadvantages |
|---|---|---|---|---|
| Shan et al. (2012) | NLM | Breast | Better accuracy with fair computational speed | Cannot segment more than one lesion as well as failed under severe shadowing effect |
| Guo et al. (2016) | Neutrosophic similarity score | Breast | Segment the tumor accurately and effectively | Semiautomatic Fewer images used |
| Zhang et al. (2010b) | NS watershed | Breast | Fully automatic, effective, and robust Segment low-contrast US images with high accuracy | Oversegmentation |
| Nugroho et al. (2017) | Normalization method | Breast | Enhance contrast | — |
| Lotfollahi et al. (2018) | NS-based active contour | Breast | Accurate Efficient | Semiautomatic |
| Xian et al. (2014) | Neutrosophic connectedness (neutro-connectedness) | Breast | Handle weak boundary leakage problem | Not fully automatic |
| Lal et al. (2018) | (IGNLM) clustering | Breast | Fully automatic, robust Produced homogeneous clustering even in the presence of shadow regions | Cannot segment multiple nodules |
| Koundal et al. (2012a) | NKUAN, NLEE | Thyroid | Effective in speckle removal | Edges and details are not preserved |
| Koundal et al. (2018) | NNTV | Thyroid | Speckle suppressed and edges preserved | Other organs yet to be explored |
| Koundal et al. (2016a) | Neutrosophic based nonconvex speckle reduction | Thyroid | Speckle noise suppressed | Edges and details are not preserved |
| Koundal et al. (2016b) | SNDRLS | Thyroid | Delineate more than one nodule effectively and accurately | Failed to delineate iso-echoic nodules |
| Koundal (2017) | Texture-based NS | Thyroid | Segment nodules of different tissues | Experiments are performed on fewer images |
| Kaur and Singh (2016) | NS-based FCM and neural network | Retinal images | Detected and classified various disease with 97.6% accuracy | Need to improve accuracy |

5 Conclusion
Neutrosophy is employed as a vital tool for removing uncertainty from ultrasound images, which are widely used for various applications in image processing. The ultrasound images are inherently fuzzy and contain uncertain information. The neutrosophic based methods can handle the uncertain information of the images effectively to achieve better results. The image is defined as an NS using three membership sets: true, indeterminate, and false. The NS-based methods can handle the indeterminacy and uncertainty of the images effectively. We will continue to study it in the foreseeable future. This work would help researchers in successfully solving CAD problems by thoroughly introducing various methods that can improve the ultrasound image quality and assist doctors in making decisions. Additionally, there are many characteristics still needed to be considered in the future to obtain better accuracy and performance. The present study recommends some challenges and directions for further exploring the area of ultrasound image analysis in detail. The classification techniques can be integrated with texture features to develop a CADx system that can be more helpful for diagnostic purposes. Moreover, research involving bigger real-time image datasets with feedback information is needed to validate the benefits of neutrosophic based methods on other tissues. The future research US images as most of the work is only done in B-mode US images.