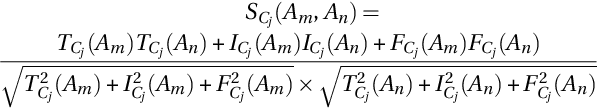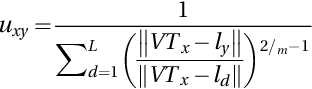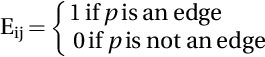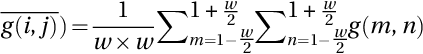A survey on neutrosophic medical image segmentation
Abdulkadir Sengur*; Umit Budak†; Yaman Akbulut‡; Murat Karabatak§; Erkan Tanyildizi§ * Department of Electrical and Electronics Engineering, Technology Faculty, Firat University, Elazig, Turkey
† Electrical and Electronics Eng. Dept., Engineering Faculty, Bitlis Eren University, Bitlis, Turkey
‡ Informatics Dept., Firat University, Elazig, Turkey
§ Department of Software Engineering, Technology Faculty, Firat University, Elazig, Turkey
Abstract
In the last decade, neutrosophic sets (NS), which are defined as the generalization of interval fuzzy sets, have become a hot topic in the computer-vision and machine-learning communities with a number of applications. Researchers in the field of computer vision have applied NS on various image-processing applications. Segmentation is a well-known process in image processing that aims to divide an input image into its regions. While the pixels in a given region should have the same property, the pixels in different regions should have different properties. In this chapter, a survey is presented on NS-based medical image segmentation. As the literature is explored, it is seen that NS-based image segmentation approaches have been applied on various medical images such as breast ultrasounds (BUS), liver computed tomography (CT), brain CTs, dermoscopy, retinal, eye angiography, dental X-rays, etc. Moreover, there have been numerous applications that have used NS in optical image segmentation. Especially, neutrosophic logic has applications in texture image segmentation. In these studies, NS has been generally used for either denoising or image enhancement. Moreover, in most studies, NS has been used for image segmentation. Besides the literature review, several well-known NS-based medical image segmentation approaches are introduced. In these methods, the NS was either used to improve the image quality by contrast enhancement and noise removal or to segment the image into regions of interest and background. The methodologies and results of the investigated methods are given in detail. The general limitations of the NS-based medical image segmentation approaches are also given. The chapter ends with some conclusions and future perspectives.
Keywords
Neutrosophic sets; Neutrosophic image; Image segmentation; Medical image analysis; Tumor detection; Lesion segmentation
1 Introduction
Image segmentation, which is quite important for computer vision, is introduced as partitioning an image into its regions-based on some criteria where the regions are meaningful and disjoint (Cheng et al., 2001). Image segmentation is generally considered an intermediate step of some pattern-recognition applications (Comaniciu, Meer, & Member, 2002). Various image-segmentation approaches have been proposed (Akbulut et al., 2018; Chen et al., 2018; Das et al., 2019; Guo et al., 2018; Jain & Laxmi, 2018; Kumar et al., 2018; Turhan et al., 2018; Wang et al., 2018). These methods are broadly classified into three categories: threshold-, edge-, and region-based methods, respectively. The threshold-based image-segmentation approaches generally use the histogram of the input image to detect single or multiple thresholds (Naidu, Rajesh Kumar, & Chiranjeevi, 2018). Edge-based image segmentation techniques aim to detect the edges in an input image. Thus, segmentation is handled by determination of the region boundaries in the input image (Zhi & Shen, 2018). Region-based image segmentation techniques initially search for some seed points in the input image and proper region growing approaches are employed to reach the boundaries of the objects. Image segmentation is also important for some medical image applications (Yang et al., 2018). In medical image analysis, highly skilled physicians spend hours to determine some regions of medical images to indicate salient regions. This procedure can be handled in seconds with a proper image segmentation approach.
In the last decade, successful applications of neutrosophy in image segmentation have appeared in the medical environment. A neutrosophic set (NS) can be seen as the generalization of fuzzy sets (Smarandache, 2003). NS is different than fuzzy sets because it uses the indeterminacy set. More specifically, in NS theory, every event is symbolized with three membership degrees: truth, falsity, and indeterminacy. In another definition, in NS, an event A is represented by its neutrality Neut A and opposite Anti A.
In this chapter, we present a survey on neutrosophic medical image segmentation. A comprehensive literature review is presented on NS-based medical image segmentation approaches. The details of the approaches, the considered medical images, and the obtained performances are investigated in the literature review. We further detail the approaches and show some visual results. The limitations and prospective studies are also investigated. The literature review is presented in Section 2. NS-based medical image segmentation approaches are introduced and their results are presented in Section 3. In Section 4, the limitations and prospective studies are described. In Section 5, we give some conclusions.
2 Literature review
The NS theory was first introduced on image segmentation by the work of Guo and Cheng (2009). Guo et al. aimed to code the gray levels of an image in the context of NS theory. Each gray level was presented with NS triplets and the indeterminacy level was reduced by applying several operations such as alpha mean and beta enhancement. Actually, these operations aimed to reduce the noise level and enhance the contrast of the input image. Then, a modified k-means clustering algorithm was used for segmentation. In other words, the segmentation was achieved by pixel clustering. Authors used some gray-level images in their experiments and the results were evaluated visually. Later, Karabatak, Guo, and Sengur (2013) presented a modified version of the NS-based image segmentation approach and applied it on color image segmentation. As the proposal of Guo and Cheng (2009) suffers from oversegmentation and fixed α and β values, Karabatak et al. (2013) proposed a novel approach to alleviate such weaknesses. The authors used an entropy-based indeterminacy member calculation to fix the oversegmentation problem and an adaptive approach was performed on the α and β parameters. Then, NS-based image segmentation attracted so many researchers from the image-processing domain, leading to dozens of studies about NS-based image segmentation being proposed so far (Akbulut et al., 2017; Gholami, Rashno, & Heshmati, 2016; Guo & Sengur, 2013; Guo & Sengur, 2015a; Guo & Sengur, 2015b; Guo & Sengur, 2015c; Guo & Şengür, 2013; Guo & Şengür, 2014; Guo, Şengür, & Tian, 2016; Jha et al., 2019; Otay & Kahraman, 2019; Sengur & Guo, 2011; Xu et al., 2018).
In the above paragraph, we briefly mentioned pioneering NS-based image segmentation studies and just cited other NS-based image segmentation approaches. So, NS-based medical image segmentation studies are reviewed in further paragraphs. Ali et al. (2018) proposed a neutrosophic orthogonal matrices-based clustering approach for image segmentation. The proposed approach initially transferred the input images into the NS domain and the inner products of the cutting matrix of the input image were calculated. Segmentation was then achieved by pixel clustering based on the orthogonal principle. The authors applied their method on a dental X-ray image dataset, which included 66 images. The authors obtained satisfactory results where the achievements were evaluated by cluster validity index values.
Ashour et al. (2018) proposed a novel image segmentation approach for dermoscopy images that was based on neutrosophic clustering and histogram estimation. The histogram estimation was used to obtain the exact number of clusters in the input image. The segmentation process was then carried out by the neutrosophic c-means clustering approach (Guo & Sengur, 2015b). The intensity and morphological features were considered in the clustering-based segmentation approach and the ISIC 2016 dermatology image dataset was used. The authors mentioned that the proposed method determined the lesion boundaries with 96.3% accuracy. Lee et al. (2018) used an NS-based approach for breast lesion segmentation in computed tomography (CT) images. The input CT images were initially converted to the neutrosophic domain and α-mean, β-enhancement, and γ-plateau operations were applied iteratively on the transferred image until the true membership value of the transferred image was no longer changed. Thus, the noises on CT images were eliminated. The RGI segmentation approach was then employed on noise-free CT images for lesion segmentation. A dataset that consisted of 122 breast lesions was used in the experiments and the average dice value was calculated for the performance evaluation metric. The presented dice value by the authors was 0.82.
Lotfollahi et al. (2018) proposed neutrosophy theory and an active contour model for breast ultrasound image segmentation. The proposed approach considered the neutrosophy theory for reducing speckle noise and enhanced the tissue-related regions in the ultrasound images. The authors also improved the active contour models by integrating a weighted region-scalable Scheme. A total of 36 breast ultrasound images were used in the experiments and 95% true positives, 6% false positives, and 90% similarity scores were presented. Anter and Hassenian (2018b) proposed a CT liver tumor segmentation approach that was based on neutrosophic theory, the clustering approach, and watershed segmentation. The proposed approach initially applied various preprocessing methods to enhance the input CT images. The enhanced CT images were transformed into the NS domain and adaptive thresholding and morphological operators were employed for further enhancing the CT images in the NS domain. The watershed process with a connected component algorithm was used on the enhanced NS images for obtaining a postsegmentation. A fast fuzzy clustering approach was considered for final segmentation of the liver tumors. A total of 105 patients’ CT slices were used in the experiments and six different indices were used to evaluate the segmentation results.
Lal, Kaur, and Gupta (2018a) proposed a neutrosophic clustering approach for tumor segmentation in B-mode breast ultrasound images. In the proposed scheme, initially the potential tumor regions were detected. Then, the desired tumor area was segmented based on the NS clustering approach that was based on the concept of information gain. The local neighborhood of each pixel was used for obtaining the information gain values. The membership values and the cluster centers were updated according to obtained values. Koundal, Gupta, and Singh (2018) proposed a fully automatic scheme for nodule segmentation in thyroid ultrasound images. The proposed approach employed an NS-based preprocessing stage for speckle reduction in thyroid ultrasound images. This process also preserved the important features that were used to determine the region of interest. The nodules were then segmented by the neutrosophic level-set method. Authors mentioned that their proposed segmentation method outperformed other methods by gaining a 95.92 ± 3.70% true positive (TP) rate. Lal, Kaur, and Gupta (2018b) developed a novel image segmentation technique that used the spatial neutrosophic clustering technique. The proposed technique extracted the boundary of tumors automatically in B-mode BUS images. The authors incorporated the spatial information into the neutrosophic ℓ-means (NLM) clustering and updated the membership values by using a type-2 membership function. A dataset that contained 60 BUS images was used in experiments and a boundary error metric was used as the performance evaluation criteria. The authors reported high performance compared with some existing methods.
Anter and Hassenian (2018a) developed an approach that was based on NS, optimization, and clustering theories. The authors used fast fuzzy c-means (FCM) and particle swarm optimization (PSO) for abdominal CT liver tumor detection. In the proposed approach, the NS was used to enhance the CT images by removing the speckle noise. Moreover, the CT images were further enhanced by removing both the high frequencies of the original images and the median filtering. The PSO was used to optimize the clustering and obtain the final segmentation of the tumors. The authors used the variance analysis, the Jaccard Index, and the dice coefficient to evaluate the segmentation results. The reported performance showed the efficiency of the proposed approach. Guo et al. (2017) proposed an efficient scheme for retinal vessel segmentation in color fundus images. The proposed method was based on the shearlet transform and NS-based indeterminacy filtering. Different from the other studies, in this work the shearlet transform was used to transfer the input fundus images into the NS domain. The indeterminacy filtering on the NS domain was used to enhance the input images. The authors used the neural network classifier for vessel pixel classification where the input features were obtained from neutrosophic images. Two publicly available datasets were used in the experimental studies and the receiver operating characteristic curve (ROC) and the area under the curve (AUC) were used for performance evaluation. The reported AUC values were 0.9476 and 0.9469 for each dataset, respectively.
Guo et al. (2013) improved a lung segmentation method with neutrosophic theory that was based on expectation maximization (EM) and binary morphological operations for pulmonary embolism detection in CT pulmonary angiography images. The developed scheme was based on an iterative NS approach. Anatomic features such as ribs and lungs were used in the initial segmentation and the segmentation was improved by using the iterative NS algorithm to obtain the final lung segmentation. In the experiments, five and 58 CT scans were used in training and test sets, respectively, and the obtained results were evaluated by various performance evaluation tests. The authors mentioned a clear improvement against the previous studies. Shan, Cheng, and Wang (2012) proposed a fully automatic breast tumor segmentation approach that was based on phase information and neutrosophic clustering. The authors initially employed preprocessing for denoising and contrast enhancement of the input CT images based on phase information, and then the region of interest was determined. After the quality of the input image was improved, the neutrosophic l-means clustering approach was used to determine tumor segmentation. The authors mentioned that the proposed neutrosophic l-means clustering approach was able to deal with uncertainty better than the other clustering approaches. Accuracy, efficiency, and sensitivity analysis were used for performance evaluation. According to the results, a true positive rate of 92.4% was presented by the authors.
Zhang (2010) proposed NS-based BUS image segmentation that was accurate, effective, and robust. The authors employed NS for developing a fully automatic algorithm. By integrating NS into their algorithm, the authors handled two conflicting opinions about speckles in ultrasound images. According to the authors, the proposed method highly improved the BUS segmentation where more accurate and robust segmentations were obtained. Xian, Cheng, and Zhang (2014) proposed an NS-based segmentation approach for BUS images. The authors developed the neutrosophic connectedness approach. The neutrosophic connectedness was used to characterize the uncertainty and indeterminacy of the spatial topological properties of the BUS images. A breast ultrasound database with 131 cases was used in experiments and the achievement was measured by similarity ratio, false positive ratio, and average Hausdroff error. The authors reported that the proposed method produced more accurate and robust segmentation than the compared methods. Gaber et al. (2015) proposed a two-phase approach for breast cancer segmentation and classification. The former phase was composed of NS-based image enhancement and fast fuzzy c-means-based clustering. The breast parenchyma regions in the thermogram images were segmented with a postsegmentation process. Support vector machines (SVM) were used for classification of breast parenchyma into normal or abnormal cases. Accuracy, precision, and recall were used for performance evaluation. The authors mentioned 100% accuracy in classification of the normal and abnormal cases.
Mohan, Krishnaveni, and Huo (2015) developed an approach for brain tumor segmentation that was based on neutrosophic c-means (NCM) clustering. The authors initially applied nonlocal NS-based Wiener filtering for image enhancement. A further fuzzy-based image enhancement was applied on the filtered images. The segmentation was achieved by the clustering enhanced image. Various evaluation metrics such as Jaccard similarity, dice coefficient, specificity, sensitivity, accuracy, false positive rate, and false negative rate were used and 100% segmentation was obtained for 20 test images. A hybrid approach was proposed by Sayed et al. (2016) for abdominal CT images. The developed approach was based on NS and the modified watershed algorithm. The authors initially used histogram equalization and median filtering to enhance the input CT images. After improving the CT images, neutrosophic memberships were calculated. The truth membership of the input CT image was used for further postprocessing. The liver region was segmented in the improved truth membership image by using the modified watershed algorithm. Performance evaluation was achieved by calculating the accuracy score and 95% overall accuracy was obtained. Alsmadi (2018) proposed an effective approach for jaw lesion segmentation in panoramic X-ray images. More specifically, a hybrid approach was considered by the author where fuzzy c-means and NS were used. The performance evaluation was carried out by calculating the area error metrics, specificity, sensitivity and similarity analyses. The author reported that the proposed method was successful in the detection of jaw lesions.
3 NS-based medical image segmentation methods
In this section, we explore several NS-based medical image segmentation approaches. Especially, the methodology and results are presented.
3.1 NSS and level set-based BUS image segmentation (Guo et al., 2016)
Factors such as speckle noise and poor quality of BUS images have greatly challenged researchers in medical image-processing applications. As a solution to this problem, NSS and level-set algorithms were used by Guo et al. (2016) for BUS image segmentation. The authors initially transferred the input images into the NS domain by using the equations that were introduced in Guo and Cheng (2009). The belongingness degree of the true tumor regions was determined by a similarity score. The segmentation of the tumor regions from the background was carried out by using the level-set method. The level-set method is a well-known image segmentation approach that is similar to the watershed algorithm.
Fig. 1 shows the flowchart of the BUS image segmentation method suggested by Cheng et al. (2001). As seen in Fig. 1, the input image goes through various stages to obtain the segmentation results. The input image is converted to the NS image by calculating the truth, falsity, and indeterminacy membership images. The NSS is calculated based on the truth, falsity, and indeterminacy memberships. The level-set method is considered to obtain the final segmentation of the NSS processed image. Their proposed method is detailed as below:

Let A = {A1, A2, … , Am} and C = {C1, C2, … , Cn} be the set of alternatives and conditions in the NS domain, respectively, and the alternative Ai under the Cj condition denotes as {TCj(Ai), ICj(Ai), FCj(Ai)}/Ai, where the TCj(Ai), ICj(Ai) and FCj(Ai) are true (T), indeterminate (I), and false (F) membership values, respectively.
The degree of similarity between the two alternatives is measured by a similarity score in the NS domain and is calculated by the following equation (Ye, 2013):
If the NS is to be described on an image Im, BP is a bright pixel set and INS is the image NS domain. When a pixel P(x, y) is transformed in the neutrosophic set domain, PNS(x, y) = {T(x, y), I(x, y), F(x, y)}, where T(x, y), I(x, y), and F(x, y) are represented as memberships of bright, indeterminate, and dark pixels set, respectively.
Later, the authors applied the level-set algorithm to the NSS output. The level-set method is a well-known image segmentation approach and readers may refer to Osher and Sethian (1988) for detailed information about the method. Level-set approach-based image segmentation methods are usually used in two different ways such as edge- or region-based methods (Chan & Vese, 2001; Kimmel et al., 1997).
From the results that were given in Guo et al. (2016), the obtained segmentation results are quite similar to the ground-truth segmentations. The boundaries of the segmented regions are uniform and do not contain any irregular shapes. The authors also compared their achievements with a hybrid approach where NS and FCM algorithms were used. As a hybrid method's achievements are considered visually, it is seen that the boundaries of the segmented region are irregular when compared with ground-truth segmentation results. In other words, the achievements of NSS and level set-based methods are more sensitive to edge responses.
3.2 BUS image segmentation based on neutrosophic l-means clustering (Shan et al., 2012)
Shan et al. (2012) developed a fully automatic breast tumor detection method that was based on neutrosophic l-means (NLM) clustering. The authors opted to improve the image contrast by using a new phase feature and a new neutrosophic clustering method was used to determine the correct lesion border. Fig. 2 shows the flowchart of Shan et al.’s proposal.

As seen in Fig. 2, the proposed method consists of four main steps: ROI generation, speckle noise reduction, contrast improvement, and NLM clustering. The BUS images generally contain many different structures such as connective tissue, fat, and muscle. Furthermore, the lesion area is smaller than the whole image. That's why; the authors considered to use a region of interest (ROI) to increase the speed and accuracy of segmentation. The ROI defined as a rectangular region (Joo et al., 2004; Yap, Edirisinghe, & Bez, 2008) was applied in two steps: automatic seed point detection and region growing. Then, the ROI image was fed into the speckle reducing anisotropic diffusion (SRAD) method, which was previously proposed by Yu, Molloy, and Acton (2004). The SRAD approach is edge-sensitive diffusion for spotted images; likewise, conventional anisotropic diffusion is edge-sensitive diffusion for ruined images with additional noise. Then, the phase in max-energy orientation (PMO), which is a robust method for boundary detection from the image (Noble & Boukerroui, 2006), was applied to the smoothed ROI image. Finally, the authors applied the NLM algorithm for detection of tumor boundaries of the BUS image. The NLM is a new clustering method based on fuzzy c-tools and neutrosophic images and consists of six steps (Shan et al., 2012):
- (1) Designate membership matrix as U(k) = [uxy] and k = 0. Where x, y, and k are pixel index, cluster index, and iteration number, respectively.
- (2) Increase iteration by k, calculate true (T(k)), indeterminate (I(k)), and false (F(k)) for each k value and transform T(k) and I(k) into vectors VT and VI.
- (3) Determine the center vectors L(k) = [lq] as:
where m and N are the membership parameter and total pixel number in the given image, respectively.
- (4) Update membership matrix as U(k + 1) = [uxy] by using:
where L shows the number of clusters.
- (5) Update image,
where w, (i, j), and λ are window size, pixel at the window center, and indeterminacy threshold, respectively.
The authors also used the FCM algorithm to compare with their method. As reported in the paper, the tumor regions determined by the NLM method are quite similar to those of the radiologist. The borders of the segmentation are uniform and do not contain any holes in the background regions. The boundaries of the segmented regions do not contain any irregular shapes.
3.3 Iterative NS-based lung segmentation in thoracic CT images (Guo et al., 2013)
The detection of the lung in CT images is important for the detection of lung diseases and lung abnormalities. Besides, probable pathologies of different sizes in CT images make it difficult to accurately detect lung regions. For this reason, Guo et al. (2013) developed a novel approach that provides accurate lung segmentation. Because the authors could not accurately detect the lung limits in patients with lung diseases by the previously developed expectation-maximization (EM) analysis and morphological operations (EMM)-based method, they thought of using an iterative NS-based lung segmentation approach to improve the EMM segmentation method using anatomical features such as ribs and lungs. First, they extracted the lung regions with their developed EMM model by using the three-dimensional (3D) hierarchical EM segmentation method. Then, the initial EEM segmentation result was fed into INLS to determine the boundaries of the final lung regions.
In addition, the authors evaluated the segmentation results according to those segmented by a radiologist manually using percentage overlap area (POA), Hausdorff distance (HD), and average distance (AD) criteria. The reported mean and standard deviation of the POA, HD, and AD values were improved from 85.4 ± 18.14%, 22.6 ± 29.4 mm, and 3.5 ± 5.4 mm to 91.2 ± 6.7%, 16.0 ± 11.3 mm, and 2.5 ± 1.0 mm by using the combination of EMM and INLS, iteratively.
3.4 Spatial NS clustering and level set-based thyroid nodule segmentation in ultrasound images (Koundal, Gupta, & Singh, 2016)
One of the well-known imaging modalities for the detection of thyroid nodules is ultrasound (US) imaging (Koundal, 2012). After detection of the thyroid nodule, it must be delineated in the US images. Delineation of thyroid nodules accurately and effectively requires expert radiologists. As doing this delineation manually is a time-consuming operation due to speckle noise and low contrast of thyroid US images, automatic delineation of thyroid nodules has become a significant case for radiologists. In this work, Koundal et al. (2016) used spatial neutrosophic clustering and the level-set method to determine the thyroid nodules in ultrasound images. The proposed method consists of three steps that are all automatically run. These steps are given in Fig. 3.

As Fig. 3 shows, the first step was the extraction of ROI. It is clear that the extraction of ROI saves time due to reducing computational costs (Savelonas et al., 2007). In the second step, US images are transformed into the NS domain. Eqs. (7)–(12) were used to calculate the NS domain image memberships. Thus, each of the pixels in the transformed image has three membership degrees: true T, false F, and indeterminacy I. Here, spatial neutrosophic L-means (SNLM) clustering combines spatial information such as blur, edge, and mean with neutrosophic L-means (NLM) clustering (Shan et al., 2012). In the NS domain, the Tij, Fij, and Iij functions are defined as the following:
where Tij, Fij, and Iij are the truth, falsity, and indeterminacy memberships in NS space, respectively. ![]() is the pixel's local mean on the window, Bij is the blur matrix, and Eij is the edge matrix. Level-set function requires the result of SNLM clustering to initialize itself. In the third step, the proposed SNDRLS method for thyroid US image segmentation is successfully completed. The proposed method is fully automatic without any human intervention.
is the pixel's local mean on the window, Bij is the blur matrix, and Eij is the edge matrix. Level-set function requires the result of SNLM clustering to initialize itself. In the third step, the proposed SNDRLS method for thyroid US image segmentation is successfully completed. The proposed method is fully automatic without any human intervention.
In this work, 42 subjects were contributed to collect B-mode thyroid US images as a dataset. Six metrics were considered to evaluate the achievement of the proposed method on thyroid US images: TP, false positive (FP), overlap metric (OM), dice coefficient (DC), the HD, and mean absolute distance (MAD).
The comparison of the proposed SNDRLS method was carried out with other methods such as neutrosophic watershed (Zhang, Zhang, & Cheng, 2010), active contour without edges (ACWE) (Chan, Yezrielev Sandberg, & Vese, 2000), fuzzy level-set method (FLSM) (Li et al., 2011), and distance regularizer level-set evolution (DRLSE) (Li et al., 2010) using seven evaluation metrics: TP, FP, OM, DC, MAD, HD and execution time on the entire dataset.
3.5 NS and modified watershed algorithm-based segmentation approach for abdominal CT Liver Parenchyma (Sayed et al., 2016)
Liver cancer is one of the most common diseases worldwide, and it is often fatal. It is important to detect the disease in the early stages and to accurately determine the stage of the cancer. Computed tomography (CT) images are generally used to diagnose liver cancer because the CT offers more detail than general X-ray equipment. The segmentation of CT images by the radiologist or other experts is very time consuming and tedious. On the other hand, automatic segmentation of CT images is a crucial and difficult task for liver segmentation. Therefore, Sayed et al. (2016) proposed a fully automated liver segmentation approach based on NS and a modified watershed algorithm. The proposed approach has three stages: (1) preprocessing, (2) transformation, and (3) postprocessing. Fig. 4 shows the architecture of the proposed NS segmentation approach.

Each stage is detailed as follows by researchers:
Preprocessing stage: Preprocessing is one of the indispensable steps of image segmentation to eliminate noise, enhance contrast, and resize images. When you remove noise from the image, at the same time, preserving the edges of the image is critical for the success of image segmentation. All images in the dataset (Mostafa et al., 2012) are JPEG format with 630 × 630. First, CT images were converted to grayscale and resized to 256 × 256. Then, the median filter was applied to remove the noise. Finally, the output of the median filter was fed into a histogram equalization filter to adjust the dynamic range and contrast of the image.
Transformation stage: The output grayscale image of the preprocessing stage was transformed into the NS domain. Thus, each pixel has three neutrosophic subsets: truth T, false F, and indeterminate I. T represents the objects, F represents the background of the image, and I represents the edges of the image. An NS image is defined as follows:
where PNS(i, j) is the pixel in NS, ![]() is the local mean of the image, and Ho(i, j) is the homogeneity value of T at (i, j) (the absolute value of difference between intensity g(i, j) and its local mean value
is the local mean of the image, and Ho(i, j) is the homogeneity value of T at (i, j) (the absolute value of difference between intensity g(i, j) and its local mean value ![]() ).
).
First, researchers assigned a gradient image G(i, j) to the NS indeterminate image I(i, j). The gradient image G(i, j) was normalized according to Eq. (18).
where Gmax is the maximum and Gmin is the minimum intensity value of G(i, j).
Postprocessing stage: In the postprocessing stage, morphological operators were applied on the NS truth image T(i, j), which was obtained from the NS domain. A traditional watershed algorithm is sensitive to noise and it has an oversegmentation issue (Grau et al., 2004). Therefore, a developed watershed algorithm is employed to segment the liver parenchyma in the CT image. The developed watershed algorithm was compared with traditional watershed algorithms that use various distance types such as Euclidean distance, city block distance, chessboard distance, and quasi-Euclidean distance. In addition, several metrics were used to evaluate the performance between the developed approach and the other methods. These metrics are the dice coefficient (DC), the Jaccard index (JI), correlation, and true positive (TP). From the presented results, it was seen that the proposed approach achieved the highest accuracy rates in all metrics except TP. The adaptive thresholding method obtained a 97.62% TP value, that is, 2.19% better than the proposed approach. In addition, the worst evaluation scores were produced by the active contour method, where a 72.60% correlation value, a 75.32% DC value, a 61.46% JI value, and a 75.90% TP value were obtained. Except for the TP score, the adaptive threshold method produced the second-worst results when compared with the other. The adaptive threshold method produced an 84.87% correlation value, an 85.84% DC value, and a 76.95% JI value. On the other hand, as mentioned, the best TP value of 97.62% was produced by the adaptive threshold method. The region growing method also produced similar scores as the adaptive threshold method, where an 83.30% correlation value, an 84.06% DC value, a 74.35% JI value, and a 92.54TP value were obtained. Region growing produces the second-best scores where an 87.17% correlation value, an 88.66% DC value, an 80.52% JI value, and an 87.64% TP value were obtained.
The performance evaluation of the proposed method was carried out on CT images from 30 different patients by using some measurements such as true positive ratio, correlation, Jaccard index, and dice coefficient. The segmentation results show that the proposed method yields better results than the most common methods.
4 Limitations of NS-based medical image segmentation approaches
As the literature review shows, it is seen that NS has attracted much attention in medical image analysis. Especially, researchers used NS-based approaches in the detection of various lesions in medical images. As successful, robust, and efficient applications of NS in medical image segmentation have been proposed, there have been some limitations of NS-based approaches, including the following:
- (1) In almost all NS-based medical image segmentation approaches, mapping the images from the intensity domain into the neutrosophic set domain has been carried out by using the equations of Guo and Cheng (2009). In Guo and Cheng (2009), the truth membership (T) was defined as the normalized local mean image and the falsity membership (F) was defined as 1 − T. The indeterminacy membership (I) was defined as the normalized absolute value of the local difference. These membership expressions were used for all medical image types. This situation can be seen as a conflict because different medical images have different structures and necessitate different membership definitions.
- (2) In almost all studies, the authors used α-mean and β-enhancement operations for making the input images more convenient for subsequent clustering procedures. These procedures were not convenient for all medical image types and need to be adjusted for different medical images. In addition, for other NS-based methods such as Neutrosophic Similarity Score and neutrosophic c-means, many parameters need to be tuned on the training set. The segmentation algorithm depends on the specific images.
- (3) As the NS is generally applied for image enhancement in an iterative manner, this procedure may blur the input image that causes a loss of detail of the input image. So, a cost function should be incorporated into the NS-based segmentation approaches for preventing the blur problem.
- (4) In almost all NS-based medical image segmentation approaches, NS has been combined with another scheme such as clustering, graph cut, level set, active contour, and mean shift. No new segmentation framework has been proposed where NS is only used for image segmentation. In other words, in all studies, the NS part is similar but the combined part is different.
5 Conclusions
In this survey, neutrosophy theory-based medical image segmentation approaches are investigated. Neutrosophy, which comes from neutrality, expresses the attributes with truth, falsity, and indeterminacy memberships. It is deduced as the extension of the fuzzy sets. Generally, neutrosophy deals with uncertainty. Uncertainty, which can be considered as noise in image processing, is challenging in medical image segmentation. This situation makes the NS strong and robust in image-processing applications.
NS-based image segmentation approaches have achieved impressive performances in medical image segmentation applications. Especially, NS is powerful in image denoising and clustering, which can be seen in the building blocks of image segmentation. The general trend in NS-based medical image segmentation is transforming the input image into the NS domain and applying some operators for making the image more convenient for segmentation. Moreover, NS-based clustering algorithms have been developed for gray level clustering for image segmentation.
The above-mentioned situation should be adjusted for different type of medical images. In other words, the NS-based methodology that is applied to ultrasound images should not be applied to CT images. Convenient methods should be developed. The fuzzy membership functions can be considered to construct truth, falsity, and indeterminacy memberships. Some rule-based approaches can be developed to construct supervised medical image segmentation approaches.
In future studies, the NS-based deep-learning methods should be developed by the researchers. Recently, some attempts can be seen where deep learning and NS have been combined in some applications. However, that works only using NS as a preprocessing tool. So, more comprehensive studies should be developed. Similar to fuzzy neural networks, deep neutrosophic neural networks can be developed. Especially, NS and deep learning-based medical image segmentation may achieve better segmentation in the next few years.