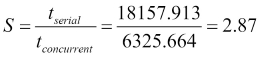The k-means clustering algorithm is a clustering algorithm to group a set of items not previously classified into a predefined number of k clusters. It's very popular within the data mining and machine learning world to organize and classify data in an unsupervised way.
Each item is normally defined by a vector of characteristics or attributes. All the items have the same number of attributes. Each cluster is also defined by a vector with the same number of attributes that represents all the items classified into that cluster. This vector is named the centroid. For example, if the items are defined by numeric vectors, the clusters are defined by the mean of the items classified into that cluster.
Basically, the algorithm has four steps:
- Initialization: In the first step, you have to create the initial vectors that represent the K clusters. Normally, you will initialize those vectors randomly.
- Assignment: Then, you classify each item into a cluster. To select the cluster, you calculate the distance between the item and every cluster. You will use a distance measure as the Euclidean distance to calculate the distance between the vector that represents the item and the vector to represents the cluster. You will assign the item to the cluster with the shortest distance.
- Update: Once all the items have been classified, you have to recalculate the vectors that define each cluster. As we mentioned earlier, you normally calculate the mean of all the vectors of the items classified into the cluster.
- End: Finally, you check whether some item has changed its assignment cluster. If there has been any change, you go to the assignment step again. Otherwise, the algorithm ends, and you have your items classified.
This algorithm has the following two main limitations:
- If you make a random initialization of the initial vectors of the clusters, as we suggested earlier, two executions to classify the same item set may give you different results.
- The numbers of cluster are previously predefined. A bad choice of this attribute will give you poor results from a classification point of view.
Despite of this, this algorithm is very popular to cluster different kinds of items. To test our algorithm, you are going to implement an application to cluster a set of documents. As a document collection, we have taken a reduced version of the Wikipedia pages with information about movies corpus we introduced in Chapter 4, Getting Data from the Tasks – The Callable and Future Interfaces. We only took 1,000 documents. To represent each document, we have to use the vector space model representation. With this representation, each document is represented as a numeric vector where each dimension of the vector represents a word or a term and its value is a metric that defines the importance of that word or term in the document.
When you represent a document collection using the vector space model, the vectors will have as many dimensions as the number of different words of the whole collection, so the vectors will have a lot of zero values because each document doesn't have all the words. You can use a more optimized representation in memory to avoid all those zero values and save memory increasing the performance of your application.
In our case, we have chosen term frequency–inverse document frequency (tf-idf) as the metric that defines the importance of each word and the 50 words with higher tf-idf as the terms that represents each document.
We use two files: the movies.words file stores a list of all the words used in the vectors, and the movies.data stores the representation of each document. The movies.data file has the following format:
10000202,rabona:23.039285705435507,1979:8.09314752937111,argentina:7.953798614698405,la:5.440565539075689,argentine:4.058577338363469,editor:3.0401515284855267,spanish:2.9692083275217134,image_size:1.3701158713905104,narrator:1.1799670194306195,budget:0.286193223652206,starring:0.25519156764102785,cast:0.2540127604060545,writer:0.23904044207902764,distributor:0.20430284744786784,cinematography:0.182583823735518,music:0.1675671228903468,caption:0.14545085918028047,runtime:0.127767002869991,country:0.12493801913495534,producer:0.12321749670640451,director:0.11592975672109682,links:0.07925582303812376,image:0.07786973207561361,external:0.07764427108746134,released:0.07447174080087617,name:0.07214163435745059,infobox:0.06151153983466272,film:0.035415118094854446
Here, 10000202 is the identifier of the document, and the rest of the file follows the formant word:tfxidf.
As with other examples, we are going to implement the serial and concurrent versions and execute both versions to verify that the Fork/Join framework gives us an improvement of the performance of this algorithm.
There are some parts that are shared between the serial and concurrent versions. These parts include:
VocabularyLoader: This is a class to load the list of words that forms the vocabulary of our corpus.Word,Document, andDocumentLoader: These three classes to load the information about the documents. These classes have a little difference between the serial and concurrent versions of the algorithm.DistanceMeasure: This is a class to calculate the Euclidean distance between two vectors.DocumentCluster: This is a class to store the information about the clusters.
Let's see these classes in detail.
As we mentioned before, our data is stored in two files. One of those files is the movies.words file. This file stores a list with all the words used in the documents. The VocabularyLoader class will transform that file into HashMap. The key of HashMap is the whole word, and the value is an integer value with the index of that word in the list. We use that index to determine the position of the word in the vector space model that represents each document.
The class has only one method, named load(), that receives the path of the file as a parameter and returns the HashMap:
public class VocabularyLoader {
public static Map<String, Integer> load (Path path) throws IOException {
int index=0;
HashMap<String, Integer> vocIndex=new HashMap<String, Integer>();
try(BufferedReader reader = Files.newBufferedReader(path)){
String line = null;
while ((line = reader.readLine()) != null) {
vocIndex.put(line,index );
index++;
}
}
return vocIndex;
}
}These classes store all the information about the documents we will use in our algorithm. First, the Word class stores information about a word in a document. It includes the index of the word and the tf-idf of that word in the document. This class only includes those attributes (int and double, respectively), and implements the Comparable interface to sort two words using their tf-idf value, so we don't include the source code of this class.
The Document class stores all the relevant information about the document. First, an array of Word objects with the words in the document. This is our representation of the vector space model. We only store the words used in the document to save a lot of memory space. Then, a String with the name of the file that stores the document and finally a DocumentCluster object to know the cluster associated with the document. It also includes a constructor to initialize those attributes and methods to get and set their value. We only include the code of the setCluster() method. In this case, this method will return a Boolean value to indicate if the new value of this attribute is the same as the old value or a new one. We will use that value to determine if we stop the algorithm or not:
public boolean setCluster(DocumentCluster cluster) {
if (this.cluster == cluster) {
return false;
} else {
this.cluster = cluster;
return true;
}
}Finally, the DocumentLoader class loads the information about the document. It includes a static method, load() that receives the path of the file, and the HashMap with the vocabulary and returns an Array of Document objects. It loads the file line by line and converts each line to a Document object. We have the following code:
public static Document[] load(Path path, Map<String, Integer> vocIndex) throws IOException{
List<Document> list = new ArrayList<Document>();
try(BufferedReader reader = Files.newBufferedReader(path)) {
String line = null;
while ((line = reader.readLine()) != null) {
Document item = processItem(line, vocIndex);
list.add(item);
}
}
Document[] ret = new Document[list.size()];
return list.toArray(ret);
}To convert a line of the text file to a Document object, we use the processItem() method:
private static Document processItem(String line,Map<String, Integer> vocIndex) {
String[] tokens = line.split(",");
int size = tokens.length - 1;
Document document = new Document(tokens[0], size);
Word[] data = document.getData();
for (int i = 1; i < tokens.length; i++) {
String[] wordInfo = tokens[i].split(":");
Word word = new Word();
word.setIndex(vocIndex.get(wordInfo[0]));
word.setTfidf(Double.parseDouble(wordInfo[1]));
data[i - 1] = word;
}
Arrays.sort(data);
return document;
}As we mentioned earlier, the first item in the line is the identifier of the document. We obtain it from tokens[0], and we pass it to the Document class constructor. Then, for the rest of the tokens, we split them again to obtain the information of every word that includes the whole word and the tf-idf value.
This class calculates the Euclidean distance between a document and a cluster (represented as a vector). The words in our word arrays after sorting are placed in the same order as in centroid array, but some words might be absent. For such words, we assume that tf-idf is zero, so the distance is just the square of the corresponding value from the centroid array:
public class DistanceMeasurer {
public static double euclideanDistance(Word[] words, double[] centroid) {
double distance = 0;
int wordIndex = 0;
for (int i = 0; i < centroid.length; i++) {
if ((wordIndex < words.length) (words[wordIndex].getIndex() == i)) {
distance += Math.pow( (words[wordIndex].getTfidf() - centroid[i]), 2);
wordIndex++;
} else {
distance += centroid[i] * centroid[i];
}
}
return Math.sqrt(distance);
}
}This class stores the information about each cluster generated by the algorithm. This information includes a list of all the documents associated with this cluster and the centroid of the vector that is the vector that represents the cluster. In this case, this vector has as many dimensions as words are in the vocabulary. The class has the two attributes, a constructor to initialize them, and methods to get and set their value. It also includes two very important methods. First, the calculateCentroid() method. It calculates the centroid of the cluster as the mean of the vectors that represents the documents associated with this cluster. We have the following code:
public void calculateCentroid() {
Arrays.fill(centroid, 0);
for (Document document : documents) {
Word vector[] = document.getData();
for (Word word : vector) {
centroid[word.getIndex()] += word.getTfidf();
}
}
for (int i = 0; i < centroid.length; i++) {
centroid[i] /= documents.size();
}
}The second method is the initialize() method that receives a Random object and initializes the centroid vector of the cluster with random numbers as follows:
public void initialize(Random random) {
for (int i = 0; i < centroid.length; i++) {
centroid[i] = random.nextDouble();
}
}Once we have described the common parts of the application, let's see how to implement the serial version of the k-means clustering algorithm. We are going to use two classes: SerialKMeans, which implements the algorithm, and SerialMain, which implements the main() method to execute the algorithm.
The SerialKMeans class implements the serial version of the k-means clustering algorithm. The main method of the class is the calculate() method. It receives the following as parameters:
- The array of
Documentobjects with the information about the documents - The number of clusters you want to generate
- The size of the vocabulary
- A seed for the random number generator
The method returns an Array of DocumentCluster object. Each cluster will have the list of documents associated with it. First, the document creates the Array of clusters determined by the numberClusters parameter and initializes them using the initialize() method and a Random object as follows:
public class SerialKMeans {
public static DocumentCluster[] calculate(Document[] documents, int clusterCount, int vocSize, int seed) {
DocumentCluster[] clusters = new DocumentCluster[clusterCount];
Random random = new Random(seed);
for (int i = 0; i < clusterCount; i++) {
clusters[i] = new DocumentCluster(vocSize);
clusters[i].initialize(random);
}Then, we repeat the assignment and update phases until all the documents stay in the same cluster. Finally, we return the array of clusters with the final organization of the documents as follows:
boolean change = true;
int numSteps = 0;
while (change) {
change = assignment(clusters, documents);
update(clusters);
numSteps++;
}
System.out.println("Number of steps: "+numSteps);
return clusters;
}The assignment phase is implemented in the assignment() method. This method receives the array of Document and DocumentCluster objects. For each document, it calculates the Euclidean distance between the document and all the clusters and assigns the document to the cluster with the lowest distance. It returns a Boolean value to indicate if one or more of the documents has changed their assigned cluster from one step to the next one. We have the following code:
private static boolean assignment(DocumentCluster[] clusters, Document[] documents) {
boolean change = false;
for (DocumentCluster cluster : clusters) {
cluster.clearClusters();
}
int numChanges = 0;
for (Document document : documents) {
double distance = Double.MAX_VALUE;
DocumentCluster selectedCluster = null;
for (DocumentCluster cluster : clusters) {
double curDistance = DistanceMeasurer.euclideanDistance(document.getData(), cluster.getCentroid());
if (curDistance < distance) {
distance = curDistance;
selectedCluster = cluster;
}
}
selectedCluster.addDocument(document);
boolean result = document.setCluster(selectedCluster);
if (result)
numChanges++;
}
System.out.println("Number of Changes: " + numChanges);
return numChanges > 0;
}The update step is implemented in the update() method. It receives the array of DocumentCluster with the information of the clusters, and it simply recalculates the centroid of each cluster:
private static void update(DocumentCluster[] clusters) {
for (DocumentCluster cluster : clusters) {
cluster.calculateCentroid();
}
}
}The SerialMain class
The SerialMain class includes the main() method to launch the tests of the k-means algorithm. First, it loads the data (words and documents) from the files:
public class SerialMain {
public static void main(String[] args) {
Path pathVoc = Paths.get("data", "movies.words");
Map<String, Integer> vocIndex=VocabularyLoader.load(pathVoc);
System.out.println("Voc Size: "+vocIndex.size());
Path pathDocs = Paths.get("data", "movies.data");
Document[] documents = DocumentLoader.load(pathDocs, vocIndex);
System.out.println("Document Size: "+documents.length);Then, it initializes the number of clusters we want to generate and the seed for the random number generator. If they don't come as parameters of the main() method, we use a default value as follows:
if (args.length != 2) {
System.err.println("Please specify K and SEED");
return;
}
int K = Integer.valueOf(args[0]);
int SEED = Integer.valueOf(args[1]);
}Finally, we launch the algorithm measuring its execution time and write the number of documents per cluster.
Date start, end;
start=new Date();
DocumentCluster[] clusters = SerialKMeans.calculate(documents, K ,vocIndex.size(), SEED);
end=new Date();
System.out.println("K: "+K+"; SEED: "+SEED);
System.out.println("Execution Time: "+(end.getTime()- start.getTime()));
System.out.println(
Arrays.stream(clusters).map (DocumentCluster::getDocumentCount).sorted (Comparator.reverseOrder())
.map(Object::toString).collect( Collectors.joining(", ", "Cluster sizes: ", "")));
}
}To implement the concurrent version of the algorithm, we have used the Fork/Join framework. We have implemented two different tasks based on the RecursiveAction class. As we mentioned earlier, the RecursiveAction task is used when you want to use the Fork/Join framework with tasks that do not return a result. We have implemented the assignment and the update phases as tasks to be executed in a Fork/Join framework.
To implement the concurrent version of the k-means algorithm, we are going to modify some of the common classes to use concurrent data structures. Then, we are going to implement the two tasks, and finally, we are going to implement the ConcurrentKMeans that implements the concurrent version of the algorithm and the ConcurrentMain class to test it.
As we mentioned earlier, we have implemented the assignment and update phases as tasks to be implemented in the Fork/Join framework.
The assignment phase assigns a document to the cluster that has the lowest Euclidean distance with the document. So, we have to process all the documents and calculate the Euclidean distances of all the documents and all the clusters. We are going to use the number of documents a task has to process as the measure to control whether we have to split the task or not. We start with the tasks that have to process all the documents and we are going to split them until we have tasks that have to process a number of documents lower than a predefined size.
The AssignmentTask class has the following attributes:
- The array of
ConcurrentDocumentClusterobjects with the data of the clusters - The array of
ConcurrentDocumentobjects with the data of the documents - Two integer attributes,
startandend, that determines the number of documents the task has to process - An
AtomicIntegerattribute,numChanges, that stores the number of documents that have changed its assigned cluster from the last execution to the current one - An integer attribute,
maxSize, that stores the maximum number of documents a task can process
We have implemented a constructor to initialize all these attributes and methods to get and set its values.
The main method of these tasks is (as with every task) the compute() method. First, we check the number of documents the tasks have to process. If it's less or equal than the maxSize attribute, we process those documents. We calculate the Euclidean distance between each document and all the clusters and select the cluster with the lowest distance. If it's necessary, we increment the numChanges atomic variable using the incrementAndGet() method. The atomic variable can be updated by more than one thread at the same time without using synchronization mechanisms and without causing any memory inconsistencies. Refer to the following code:
protected void compute() {
if (end - start <= maxSize) {
for (int i = start; i < end; i++) {
ConcurrentDocument document = documents[i];
double distance = Double.MAX_VALUE;
ConcurrentDocumentCluster selectedCluster = null;
for (ConcurrentDocumentCluster cluster : clusters) {
double curDistance = DistanceMeasurer.euclideanDistance (document.getData(), cluster.getCentroid());
if (curDistance < distance) {
distance = curDistance;
selectedCluster = cluster;
}
}
selectedCluster.addDocument(document);
boolean result = document.setCluster(selectedCluster);
if (result) {
numChanges.incrementAndGet();
}
}If the number of documents the task has to process is too big, we split that set into two parts and create two new tasks to process each of those parts as follows:
} else {
int mid = (start + end) / 2;
AssignmentTask task1 = new AssignmentTask(clusters, documents, start, mid, numChanges, maxSize);
AssignmentTask task2 = new AssignmentTask(clusters, documents, mid, end, numChanges, maxSize);
invokeAll(task1, task2);
}
}To execute those tasks in the Fork/Join pool, we have used the invokeAll() method. This method will return when the tasks have finished their execution.
The update phase recalculates the centroid of each cluster as the mean of all the documents. So, we have to process all the clusters. We are going to use the number of clusters a task has to process as the measure to control if we have to split the task or not. We start with a task that has to process all the clusters, and we are going to split it until we have tasks that have to process a number of clusters lower than a predefined size.
The UpdateTask class has the following attributes:
- The array of
ConcurrentDocumentClusterobjects with the data of the clusters - Two integer attributes,
startandend, that determine the number of clusters the task has to process - An integer attribute,
maxSize, that stores the maximum number of clusters a task can process
We have implemented a constructor to initialize all these attributes and methods to get and set its values.
The compute() method first checks the number of clusters the task has to process. If that number is less than or equal to the maxSize attribute, it processes those clusters and updates their centroid:
@Override
protected void compute() {
if (end - start <= maxSize) {
for (int i = start; i < end; i++) {
ConcurrentDocumentCluster cluster = clusters[i];
cluster.calculateCentroid();
}If the number of clusters the task has to process is too big, we are going to divide the set of clusters the task has to process in two and create two tasks to process each of that part as follows:
} else {
int mid = (start + end) / 2;
UpdateTask task1 = new UpdateTask(clusters, start, mid, maxSize);
UpdateTask task2 = new UpdateTask(clusters, mid, end, maxSize);
invokeAll(task1, task2);
}
}The ConcurrentKMeans class implements the concurrent version of the k-means clustering algorithm. As the serial version, the main method of the class is the calculate() method. It receives the following as parameters:
The calculate() method returns an array of the ConcurrentDocumentCluster objects with the information of the clusters. Each cluster has the list of documents associated with it. First, the document creates the array of clusters determined by the numberClusters parameter and initializes them using the initialize() method and a Random object:
public class ConcurrentKMeans {
public static ConcurrentDocumentCluster[] calculate(ConcurrentDocument[] documents int numberCluster int vocSize, int seed, int maxSize) {
ConcurrentDocumentCluster[] clusters = new ConcurrentDocumentCluster[numberClusters];
Random random = new Random(seed);
for (int i = 0; i < numberClusters; i++) {
clusters[i] = new ConcurrentDocumentCluster(vocSize);
clusters[i].initialize(random);
}Then, we repeat the assignment and update phases until all the documents stay in the same cluster. Before the loop, we create ForkJoinPool that is going to execute that task and all of its subtasks. Once the loop has finished, as with other Executor objects, we have to use the shutdown() method with a Fork/Join pool to finish its executions. Finally, we return the array of clusters with the final organization of the documents:
boolean change = true;
ForkJoinPool pool = new ForkJoinPool();
int numSteps = 0;
while (change) {
change = assignment(clusters, documents, maxSize, pool);
update(clusters, maxSize, pool);
numSteps++;
}
pool.shutdown();
System.out.println("Number of steps: "+numSteps); return clusters;
}The assignment phase is implemented in the assignment() method. This method receives the array of clusters, the array of documents, and the maxSize attribute. First, we delete the list of associated documents to all the clusters:
private static boolean assignment(ConcurrentDocumentCluster[] clusters, ConcurrentDocument[] documents, int maxSize, ForkJoinPool pool) {
boolean change = false;
for (ConcurrentDocumentCluster cluster : clusters) {
cluster.clearDocuments();
}Then, we initialize the necessary objects: an AtomicInteger to store the number of documents whose assigned cluster has changed and the AssignmentTask that will begin the process.
AtomicInteger numChanges = new AtomicInteger(0);
AssignmentTask task = new AssignmentTask(clusters, documents, 0, documents.length, numChanges, maxSize);
Then, we execute the tasks in the pool in an asynchronous way using the execute() method of ForkJoinPool and wait for its finalization with the join() method of the AssignmentTask object as follows:
pool.execute(task);
task.join();Finally, we check the number of documents that has changed its assigned cluster. If there have been changes, we return the true value. Otherwise, we return the false value. We have the following code:
System.out.println("Number of Changes: " + numChanges);
return numChanges.get() > 0;
}The update phase is implemented in the update() method. It receives the array of clusters and the maxSize parameters. First, we create an UpdateTask object to update all the clusters. Then, we execute that task in the ForkJoinPool object the method receives as parameter as follows:
private static void update(ConcurrentDocumentCluster[] clusters, int maxSize, ForkJoinPool pool) {
UpdateTask task = new UpdateTask(clusters, 0, clusters.length, maxSize, ForkJoinPool pool);
pool.execute(task);
task.join();
}
}To compare the two solutions, we have executed different experiments changing the values of three different parameters:
- The k-parameter will establish the number of clusters we want to generate. We have tested the algorithms with the values 5, 10, 15, and 20.
- The seed for the
Randomnumber generator. This seed determines how the initial centroid positions. We have tested the algorithms with the values 1 and 13. - For the concurrent algorithm, the
maxSizeparameter that determines the maximum number of items (documents or clusters), a task can process without being split into other tasks. We have tested the algorithms with the values 1, 20, and 400.
We have executed the experiments using the JMH framework (http://openjdk.java.net/projects/code-tools/jmh/) that allows you to implement micro benchmarks in Java. Using a framework for benchmarking is a better solution that simply measures time using methods such as currentTimeMillis() or nanoTime(). We have executed them 10 times in a computer with a four-core processor and calculated the medium execution time of those 10 times. These are the execution times we have obtained in milliseconds:
|
Serial |
Concurrent | ||||
|---|---|---|---|---|---|
|
K |
Seed |
MaxSize=1 |
MaxSize=20 |
maxSize=400 | |
|
5 |
1 |
6676.141 |
4696.414 |
3291.397 |
3179.673 |
|
10 |
1 |
6780.088 |
3365.731 |
2970.056 |
2825.488 |
|
15 |
1 |
12936.178 |
5308.734 |
4737.329 |
4490.443 |
|
20 |
1 |
19824.729 |
7937.820 |
7347.445 |
6848.873 |
|
5 |
13 |
3738.869 |
2714.325 |
1984.152 |
1916.053 |
|
10 |
13 |
9567.416 |
4693.164 |
3892.526 |
3739.129 |
|
15 |
13 |
12427.589 |
5598.996 |
4735.518 |
4468.721 |
|
20 |
13 |
18157.913 |
7285.565 |
6671.283 |
6325.664 |
We can draw the following conclusions:
- The seed has an important and unpredictable impact in the execution time. Sometimes, the execution times are lower with seed 13, but other times are lower with seed 1.
- When you increment the number of clusters, the execution time increments too.
- The
maxSizeparameter doesn't have much influence in the execution time. The parameter K or seed has a higher influence in the execution time. If you increase the value of the parameter, you will obtain better performance. The difference is bigger between 1 and 20 than between 20 and 400. - In all the cases, the concurrent version of the algorithm has better performance than the serial one.
For example, if we compare the serial algorithm with parameters K=20 and seed=13 with the concurrent version with parameters K=20, seed=13, and maxSize=400 using the speed-up, we obtain the following result: