Also called algorithmic geometry, the applications of this field are plenty. In robotics, it is used to solve visibility problems, and motion planning, for instance. Similar applications are employed to design route planning or search algorithms in geographic information systems (GIS).
Let's describe the different categories of problems, putting emphasis on the tools to solve them, which are available in the SciPy stack.
The fundamental problems in this category are the following:
- Convex hulls: Given a set of points in space, find the smallest convex polyhedron containing them.
- Voronoi diagrams: Given a set of points in space (the seeds), compute a partition in regions consisting of all points closer to each seed.
- Triangulations: Partition the plane with triangles in a way that two triangles are either disjointed, or otherwise they share an edge or a vertex. There are different triangulations depending on the input objects or constraints on the properties of the triangles.
- Shortest paths: Given a set of obstacles in a space and two points, find the shortest path between the points that does not intersect any of the obstacles.
Tip
The problems of computation of convex hulls, basic triangulations, and Voronoi diagrams are intimately linked. The theory that explains this beautiful topic is explained in detail in a monograph in computer science titled Computational Geometry, written by Franco Preparata and Michael Shamos. It was published by Springer-Verlag in 1985.
While it is possible to compute the convex hull of a reasonably large set of points in the plane through the geometry module of the library SymPy, this is not recommended. A much faster and reliable code is available in the module scipy.spatial through the class ConvexHull, which implements a wrapper to the routine qconvex, from the Qhull libraries (http://www.qhull.org/). This routine also allows the computation of convex hulls in higher dimensions. Let's compare both methods, with the famous Lake Superior polygon, superior.poly.
Tip
The poly files represent planar straight line graphs—a simple list of vertices and edges, together with information about holes and concavities, in some cases. The running example can be downloaded from https://github.com/blancosilva/Mastering-Scipy/blob/master/chapter6/superior.poly.
This contains a polygonal description of the coastline of Lake Superior, with 7 holes (for the islands), 518 vertices, and 518 edges.
For a complete description of the poly format, refer to http://www.cs.cmu.edu/~quake/triangle.poly.html. With that information, we can write a simple reader without much effort.
Following is an example.
# part of file chapter6.py from numpy import array def read_poly(file_name): """ Simple poly-file reader, that creates a python dictionary with information about vertices, edges and holes. It assumes that vertices have no attributes or boundary markers. It assumes that edges have no boundary markers. No regional attributes or area constraints are parsed. """ output = {'vertices': None, 'holes': None, 'segments': None} # open file and store lines in a list file = open(file_name, 'r') lines = file.readlines() file.close() lines = [x.strip(' ').split() for x in lines] # Store vertices vertices= [] N_vertices,dimension,attr,bdry_markers = [int(x) for x in lines[0]] # We assume attr = bdrt_markers = 0 for k in range(N_vertices): label,x,y = [items for items in lines[k+1]] vertices.append([float(x), float(y)]) output['vertices']=array(vertices) # Store segments segments = [] N_segments,bdry_markers = [int(x) for x in lines[N_vertices+1]] for k in range(N_segments): label,pointer_1,pointer_2 = [items for items in lines[N_vertices+k+2]] segments.append([int(pointer_1)-1, int(pointer_2)-1]) output['segments'] = array(segments) # Store holes N_holes = int(lines[N_segments+N_vertices+2][0]) holes = [] for k in range(N_holes): label,x,y = [items for items in lines[N_segments + N_vertices + 3 + k]] holes.append([float(x), float(y)]) output['holes'] = array(holes) return output
Notice that loading each vertex as Point, as well as computing the convex hull with that structure, requires far too many resources and too much time. Note the difference:
In [1]: import numpy as np, matplotlib.pyplot as plt; ...: from sympy.geometry import Point, convex_hull; ...: from scipy.spatial import ConvexHull; ...: from chapter6 import read_poly In [2]: lake_superior = read_poly("superior.poly"); ...: vertices_ls = lake_superior['vertices'] In [3]: %time hull = ConvexHull(vertices_ls) CPU times: user 1.59 ms, sys: 372 µs, total: 1.96 ms Wall time: 1.46 ms In [4]: vertices_sympy = [Point(x) for x in vertices_ls] In [5]: %time convex_hull(*vertices_sympy) CPU times: user 168 ms, sys: 54.5 ms, total: 223 ms Wall time: 180 ms Out[5]: Polygon(Point(1/10, -629607/1000000), Point(102293/1000000, -635353/1000000), Point(2773/25000, -643967/1000000), Point(222987/1000000, -665233/1000000), Point(8283/12500, -34727/50000), Point(886787/1000000, -1373/2000), Point(890227/1000000, -6819/10000), Point(9/10, -30819/50000), Point(842533/1000000, -458913/1000000), Point(683333/1000000, -17141/50000), Point(16911/25000, -340427/1000000), Point(654027/1000000, -333047/1000000), Point(522413/1000000, -15273/50000), Point(498853/1000000, -307193/1000000), Point(5977/25000, -25733/50000), Point(273/2500, -619833/1000000))
Let us produce a diagram with the solution, using the computations of scipy.spatial.ConvexHull:
In [5]: plt.figure(); ...: plt.xlim(vertices_ls[:,0].min()-0.01, vertices_ls[:,0].max()+0.01); ...: plt.ylim(vertices_ls[:,1].min()-0.01, vertices_ls[:,1].max()+0.01); ...: plt.axis('off'); ...: plt.axes().set_aspect('equal'); ...: plt.plot(vertices_ls[:,0], vertices_ls[:,1], 'b.') Out[5]: [<matplotlib.lines.Line2D at 0x10ee3ab10>] In [6]: for simplex in hull.simplices: ...: plt.plot(vertices_ls[simplex, 0], ...: vertices_ls[simplex, 1], 'r-') In [7]: plt.show()
This plots the following image:

To modify the output of ConvexHull, we are allowed to pass all required qconvex controls through the parameter qhull_options. For a list of all qconvex controls and other output options, consult the Qhull manual at http://www.qhull.org/html/index.htm. In this chapter, we are content with showing the results obtained with the default controls qhull_options='Qx Qt' if the dimension of the points is greater than four, and qhull_options='Qt' otherwise.
Let's now illustrate a few advanced uses of ConvexHull. First, the computation of the convex hull of a random set of points in the 3D space. For visualization, we will use the mayavi libraries:
In [8]: points = np.random.rand(320, 3) In [9]: hull = ConvexHull(points) In [10]: X = hull.points[:, 0]; ....: Y = hull.points[:, 1]; ....: Z = hull.points[:, 2] In [11]: from mayavi import mlab In [12]: mlab.triangular_mesh(X, Y, X, hull.simplices, ....: colormap='gray', opacity=0.5, ....: representation='wireframe')
This plots the following image:

Computing the Voronoi diagram of a set of vertices (our seeds) can be done with the class Voronoi (and its companion voronoi_plot_2d for visualization) from the module scipy.spatial. This class implements a wrapper to the routine qvoronoi from the Qhull libraries, with the following default controls qhull_option='Qbb Qc Qz Qx' if the dimension of the points is greater than four, and qhull_options='Qbb Qc Qz' otherwise. For the computation of the furthest-site Voronoi diagram, instead of the nearest-site, we would add the extra control 'Qu'.
Let's work a simple example with the usual Voronoi diagram:
In [13]: from scipy.spatial import Voronoi, voronoi_plot_2d In [14]: vor = Voronoi(vertices_ls)
To understand the output, it is very illustrative to replicate the diagram that we obtain by restricting the visualization obtained by voronoi_plot_2d in a small window, centered somewhere along the north shore of Lake Superior:
In [15]: ax = plt.subplot(111, aspect='equal'); ....: voronoi_plot_2d(vor, ax=ax); ....: plt.xlim( 0.45, 0.50); ....: plt.ylim(-0.40, -0.35); ....: plt.show()
This plots the following image:

- The small dots are the original seeds with x coordinates between
0.45and0.50, and y coordinates between-0.40and-0.35. We access those values either from the original listvertices_lsor fromvor.points. - The plane gets partitioned into different regions (the Voronoi cells), one for each seed. These regions contain all the points in the plane which are closest to its seed. Each region receives an index, which is not necessarily the same index as the index of its seed in the
vor.pointslist. To access the corresponding region to a given seed, we usevor.point_region:In [16]: vor.point_region Out[16]: array([ 0, 22, 24, 21, 92, 89, 91, 98, 97, 26, 218, 219, 220, 217, 336, 224, 334, 332, 335, 324, 226, 231, 230, 453, 500, 454, 235, 234, 333, 236, 341, 340, 93, ... 199, 81, 18, 17, 205, 290, 77, 503, 469, 473, 443, 373, 376, 366, 370, 369, 210, 251, 367, 368, 377, 472, 504, 506, 502, 354, 353, 54, 42, 43, 350, 417, 414, 415, 418, 419, 425])
- Each Voronoi cell is defined by its delimiting vertices and edges (also known as ridges in Voronoi jargon). The list with the coordinates of the computed vertices of the Voronoi diagram can be obtained with
vor.vertices. These vertices were represented as bigger dots in the previous image, and are easily identifiable because they are always at the intersection of at least two edges—while the seeds have no incoming edges:In [17]: vor.vertices Out[17]: array([[ 0.88382749, -0.23508215], [ 0.10607886, -0.63051169], [ 0.03091439, -0.55536174], ..., [ 0.49834202, -0.62265786], [ 0.50247159, -0.61971784], [ 0.5028735 , -0.62003065]])
- For each of the regions, we can access the set of delimiting vertices with
vor.regions. For instance, to obtain the coordinates of the vertices that delimit the region around the fourth seed, we could issue the following command:In [18]: [vor.vertices[x] for x in vor.regions[vor.point_region[4]]] Out[18]: [array([ 0.13930793, -0.81205929]), array([ 0.11638 , -0.92111088]), array([ 0.11638 , -0.63657789]), array([ 0.11862537, -0.6303235 ]), array([ 0.12364332, -0.62893576]), array([ 0.12405738, -0.62891987])]
Care must be taken with the previous step—some of the vertices of the Voronoi cells are not actual vertices, but lie at infinity. When this is the case, they are identified with the index
-1. In this situation, to provide an accurate representation of a ridge of these characteristics, we must use the knowledge of the two seeds whose contiguous Voronoi cells intersect on said ridge—since the ridge is perpendicular to the segment defined by those two seeds. We obtain the information about those seeds withvor.ridge_points:In [19]: vor.ridge_points Out[19]: array([[ 0, 1], [ 0, 433], [ 0, 434], ..., [124, 118], [118, 119], [119, 122]])
The first entry of
vor.ridge_pointscan be read as, there is a ridge perpendicular to both the first and second seeds.
There are other attributes of the object vor that we could use to inquire properties of the Voronoi diagram, but the ones we have described should be enough to replicate the previous diagram. We leave this as a nice exercise:
- Gather the indices of the seeds from
vor.pointsthat have their x coordinates and y coordinates in the required window. Plot them. - For each of those seeds, gather information about the vertices of their corresponding Voronoi cells. Plot those vertices that are not at infinity with a different style to the seeds.
- Gather information about the ridges of each relevant region, and plot them as simple thin segments. Some of the ridges cannot be represented by their two vertices. In that case, we use the information about the seeds that determine them.
A triangulation of a set of vertices in the plane is a division of the convex hull of the vertices into triangles, satisfying one important condition. Any two given triangles can be either one of the following:
- They must be disjoint
- They must intersect only at one common vertex
- They must share one common edge
These plain triangulations have not much computational value, since some of their triangles might be too skinny—this leads to uncomfortable rounding errors, computation or erroneous areas, centers, and so on. Among all possible triangulations, we always seek one where the properties of the triangles are somehow balanced.
With this purpose in mind, we have the Delaunay triangulation of a set of vertices. This triangulation satisfies an extra condition—none of the vertices lie in the interior of the circumcircle of any triangle. We refer to triangles with this property as Delaunay triangles.
For this simpler setting, in the module scipy.spatial, we have the class Delaunay, which implements a wrapper to the routine qdelaunay from the Qhull libraries, with the controls set exactly, as in the case of the Voronoi diagram:
In [20]: from scipy.spatial import Delaunay In [21]: tri = Delaunay(vertices_ls) In [22]: plt.figure() ....: plt.xlim(vertices_ls[:,0].min()-0.01, vertices_ls[:,0].max()+0.01); ....: plt.ylim(vertices_ls[:,1].min()-0.01,vertices_ls[:,1].max()+0.01); ....: plt.axes().set_aspect('equal'); ....: plt.axis('off'); ....: plt.triplot(vertices_ls[:,0], vertices_ls[:,1], tri.simplices, 'k-'); ....: plt.plot(vertices_ls[:,0], vertices_ls[:,1], 'r.'); ....: plt.show()
This plots the following diagram:

It is possible to generate triangulations with imposed edges too. Given a collection of vertices and edges, a constrained Delaunay triangulation is a division of the space into triangles with those prescribed features. The triangles in this triangulation are not necessarily Delaunay.
We can accomplish this extra condition sometimes by subdividing each of the imposed edges. We call this triangulation conforming Delaunay, and the new (artificial) vertices needed to subdivide the edges are called Steiner points.
A constrained conforming Delaunay triangulation of an imposed set of vertices and edges satisfies a few more conditions, usually setting thresholds on the values of angles or areas of the triangles. This is achieved by introducing a new set of Steiner points, which are allowed anywhere, not only on edges.
Tip
To achieve these high-level triangulations, we need to step outside of the SciPy stack. We have a Python wrapper for the amazing implementation of mesh generators, triangle, by Richard Shewchuck (http://www.cs.cmu.edu/~quake/triangle.html). This wrapper, together with examples and other related functions, can be installed from prompt by issuing either easy_install triangle, or pip install triangle. For more information on this module, refer to the documentation online from its author, Dzhelil Rufat, at http://dzhelil.info/triangle/index.html.
Let's compute those different triangulations for our running example. We once again use the poly file with the features of Lake Superior, which we read into a dictionary with all the information about vertices, segments, and holes. The first example is that of the constrained Delaunay triangulation (cndt). We accomplish this task with the flag p (indicating that the source is a planar straight line graph, rather than a set of vertices):
In [23]: from triangle import triangulate, plot as tplot In [24]: cndt = triangulate(lake_superior, 'p') In [25]: ax = plt.subplot(111, aspect='equal'); ....: tplot.plot(ax, **cndt); ....: plt.show()
Note the improvement with respect to the previous diagram, as well as the absence of triangles outside of the original polygon:

The next step is the computation of a conforming Delaunay triangulation (cfdt). We enforce Steiner points on some segments to ensure as many Delaunay triangles as possible. We achieve this with the extra flag D:
In [26]: cfdt = triangulate(lake_superior, 'pD')
Either slight or no improvement, with respect to the previous diagram, can be observed in this case. The real improvement arises when we further impose constraints in the values of minimum angles on triangles (with the flag q) or in the maximum values of the areas of triangles (with the flag a). For instance, if we require a constrained conforming Delaunay triangulation (cncfdt) in which all triangles have a minimum angle of at least 20 degrees, we issue the following command:
In [27]: cncfq20dt = triangulate(lake_superior, 'pq20D') In [28]: ax = plt.subplot(111, aspect='equal'); ....: tplot.plot(ax, **cncfq20dt); ....: plt.show()
This presents us with a much more improved result, as seen in the following diagram:
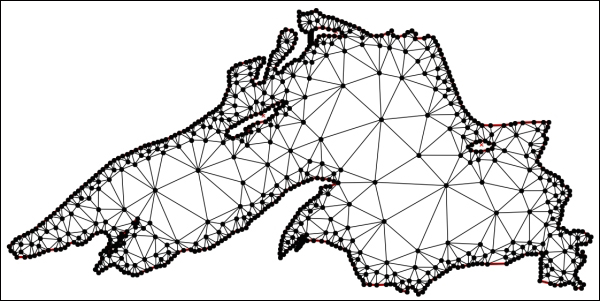
To conclude this section, we present a last example where we further impose a maximum area on triangles:
In [29]: cncfq20adt = triangulate(lake_superior, 'pq20a.001D') In [30]: ax = plt.subplot(111, aspect='equal'); ....: tplot.plot(ax, **cncfq20adt); ....: plt.show()
The last (very satisfying) diagram is as follows:

We will use the previous example to introduce a special setting to the problem of shortest paths. We pick a location on the Northwest coast of the lake (say, the vertex indexed as 370 in the original poly file), and the goal is to compute the shortest path to the furthest Southeast location on the shore, at the bottom-right corner—this is the vertex indexed as 179 in the original poly file. By a path in this setting, we mean a chain of edges of the triangulation.
In the SciPy stack, we accomplish the computation of the shortest paths on a triangulation (and in some other similar geometries that can be coded by means of graphs) by relying on two modules:
scipy.sparseis used to store a weighted-adjacency matrixGrepresenting the triangulation. Each nonzero entryG[i,j]of this adjacency matrix is precisely the length of the edge from vertexito vertexj.scipy.sparse.csgraphis the module that deals with compressed sparse graphs. This module contains routines to analyze, extract information, or manipulate graphs. Among these routines, we have several different algorithms to compute the shortest paths on a graph.Tip
For more information on the module
scipy.sparse.csgraph, refer to the online documentation at http://docs.scipy.org/doc/scipy/reference/sparse.csgraph.html.For the theory and applications of Graph Theory, one of the best sources is the introductory book by Reinhard Diestel, Graph Theory, published by Springer-Verlag.
Let's illustrate this example with proper code. We start by collecting the indices of the vertices of all segments in the triangulation, and the lengths of these segments.
In [31]: X = cncfq20adt['triangles'][:,0]; ....: Y = cncfq20adt['triangles'][:,1]; ....: Z = cncfq20adt['triangles'][:,2] In [32]: Xvert = [cncfq20adt['vertices'][x] for x in X]; ....: Yvert = [cncfq20adt['vertices'][y] for y in Y]; ....: Zvert = [cncfq20adt['vertices'][z] for z in Z] In [33]: from scipy.spatial import minkowski_distance In [34]: lengthsXY = minkowski_distance(Xvert, Yvert); ....: lengthsXZ = minkowski_distance(Xvert, Zvert); ....: lengthsYZ = minkowski_distance(Yvert, Zvert)
We now create the weighted-adjacency matrix, which we store as a lil_matrix, and compute the shortest path between the requested vertices. We gather, in a list, all the vertices included in the computed path, and plot the resulting chain overlaid on the triangulation.
Tip
A word of warning:
The adjacency matrix we are about to compute is not the distance matrix. In the distance matrix A, we include, on each entry A[i, j], the distance between any vertex i to any vertex j, regardless of being connected by an edge or not. If this distance matrix is desired, the most reliable way to compute it is by means of the routine distance_matrix in the module scipy.spatial:
>>> from scipy.spatial import distance_matrix >>> A = distance_matrix(cncfq20adt['vertices'], cncfq20adt['vertices'])
In [35]: from scipy.sparse import lil_matrix; ....: from scipy.sparse.csgraph import shortest_path In [36]: nvert = len(cncfq20adt['vertices']); ....: G = lil_matrix((nvert, nvert)) In [37]: for k in range(len(X)): ....: G[X[k], Y[k]] = G[Y[k], X[k]] = lengthsXY[k] ....: G[X[k], Z[k]] = G[Z[k], X[k]] = lengthsXZ[k] ....: G[Y[k], Z[k]] = G[Z[k], Y[k]] = lengthsYZ[k] ....: In [38]: dist_mat, pred = shortest_path(G, return_predecessors=True, ....: directed=True, ....: unweighted=False) In [39]: index = 370; ....: path = [370] In [40]: while index != 197: ....: index = pred[197, index] ....: path.append(index) ....: In [41]: print path [380, 379, 549, 702, 551, 628, 467, 468, 469, 470, 632, 744, 764, 799, 800, 791, 790, 789, 801, 732, 725, 570, 647, 177, 178, 179, 180, 181, 182, 644, 571, 201, 200, 199, 197] In [42]: ax = plt.subplot(111, aspect='equal'); ....: tplot.plot(ax, **cncfq20adt) In [43]: Xs = [cncfq20adt['vertices'][x][0] for x in path]; ....: Ys = [cncfq20adt['vertices'][x][1] for x in path] In [44]: ax.plot(Xs, Ys '-', linewidth=5, color='blue'); ....: plt.show()
This gives the following diagram:

The fundamental problems in this category are as follows:
- Point location
- Nearest neighbor
- Range searching
The problem of point location is fundamental in computational geometry, given a partition of the space into disjoint regions, we need to query the region that contains a target location.
The most basic point location problems are those where the partition is given by a single geometric object—a circle or a polygon, for example. For those simple objects that have been constructed through any of the classes in the module sympy.geometry, we have two useful methods: .encloses_point and .encloses.
The former checks whether a point is interior to a source object (but not on the border), while the latter checks whether another target object has all its defining entities in the interior of the source object:
In [1]: from sympy.geometry import Point, Circle, Triangle In [2]: P1 = Point(0, 0); ...: P2 = Point(1, 0); ...: P3 = Point(-1, 0); ...: P4 = Point(0, 1) In [3]: C = Circle(P2, P3, P4); ...: T = Triangle(P1, P2, P3) In [4]: print "Is P1 inside of C?", C.encloses_point(P1) Is P1 inside of C? True In [5]: print "Is T inside of C?", C.encloses(T) Is T inside of C? False
Of special importance is this simple setting where the source object is a polygon. The routines in the sympy.geometry module get the job done, but at the cost of too many resources and too much time. A much faster way to approach this problem is by using the Path class from the libraries of matplotlib.path. Let's see how with a quick session. First, we create a representation of a polygon as a Path:
Tip
For information about the class Path and its usage within the matplotlib libraries, refer to the official documentation at http://matplotlib.org/api/path_api.html#matplotlib.path.Path, as well as the tutorial at http://matplotlib.org/users/path_tutorial.html.
In [6]: import numpy as np, matplotlib.pyplot as plt; ...: from matplotlib.path import Path; ...: from chapter6 import read_poly; ...: from scipy.spatial import ConvexHull In [7]: superior = read_poly("superior.poly") In [8]: hull = ConvexHull(superior['vertices']) In [9]: my_polygon = Path([hull.points[x] for x in hull.vertices])
We can now ask whether a point (respectively, a sequence of points) is interior to the polygon. We accomplish this with either contains_point or contains_points:
In [10]: X = .25 * np.random.randn(100) + .5; ....: Y = .25 * np.random.randn(100) - .5 In [11]: my_polygon.contains_points([[X[k], Y[k]] for k in range(len(X))]) Out[11]: array([False, False, True, False, True, False, False, False, True, False, False, False, True, False, True, False, False, False, True, False, True, True, False, False, False, False, False, ... True, False, True, False, False, False, False, False, True, True, False, True, True, True, False, False, False, False, False], dtype=bool)
More challenging point location problems arise when our space is partitioned by a complex structure. For instance, once a triangulation has been computed, and a random location is considered, we need to query for the triangle where our target location lies. In the module scipy.spatial, we have handy routines to perform this task over Delaunay triangulations created with scipy.spatial.Delaunay. In the following example, we track the triangles that contain a set of 100 random points in the domain:
In [12]: from scipy.spatial import Delaunay, tsearch In [13]: tri = Delaunay(superior['vertices']) In [14]: points = zip(X, Y) In [15]: print tsearch(tri, points) [ -1 687 -1 647 -1 -1 -1 -1 805 520 460 647 580 -1 -1 -1 -1 304 -1 -1 -1 -1 108 723 -1 -1 -1 -1 -1 -1 -1 144 454 -1 -1 -1 174 257 -1 -1 -1 -1 -1 52 -1 -1 985 -1 263 -1 647 -1 314 -1 -1 104 144 -1 -1 -1 -1 348 -1 368 -1 -1 -1 988 -1 -1 -1 348 614 -1 -1 -1 -1 -1 -1 -1 114 -1 -1 684 -1 537 174 161 647 702 594 687 104 -1 144 -1 -1 -1 684 -1]
Tip
The same result is obtained with the method .find_simplex of the Delaunay object tri:
In [16]: print tri.find_simplex(points) [ -1 687 -1 647 -1 -1 -1 -1 805 520 460 647 580 -1 -1 -1 -1 304 -1 -1 -1 -1 108 723 -1 -1 -1 -1 -1 -1 -1 144 454 -1 -1 -1 174 257 -1 -1 -1 -1 -1 52 -1 -1 985 -1 263 -1 647 -1 314 -1 -1 104 144 -1 -1 -1 -1 348 -1 368 -1 -1 -1 988 -1 -1 -1 348 614 -1 -1 -1 -1 -1 -1 -1 114 -1 -1 684 -1 537 174 161 647 702 594 687 104 -1 144 -1 -1 -1 684 -1]
Note that, when a triangle is found, the routine reports its corresponding index in tri.simplices. If no triangle is found (which means the point is exterior to the convex hull of the triangulation), the index reported is -1.
The problem of finding the Voronoi cell that contains a given location is equivalent to the search for the nearest neighbor in a set of seeds. We can always perform this search with a brute-force algorithm—and this is acceptable in some cases—but in general, there are more elegant and less complex approaches to this problem. The key lies in the concept of k-d trees—a special case of binary space partitioning structures for organizing points, conductive to fast searches.
In the SciPy stack, we have an implementation of k-d trees; the Python class KDTree, in the module scipy.spatial. This implementation is based on ideas published in 1999 by Maneewongvatana and Mount. It is initialized with the location of our input points. Once created, it can be manipulated and queried with the following methods and attributes:
- The methods are as follows:
data: This presents the input.leafsize: This is the number of points at which the algorithm switches to brute-force. This value can be optionally offered in the initialization of theKDTreeobject.m: This is the dimension of the space where the points are located.n: This is the number of input points.maxes: This indicates the highest values of each of the coordinates of the input points.mins: This indicates the lowest values of each of the coordinates of the input points.
- The attributes are as follows:
query(self, Q, p=2.0): This is the attribute that searches for the nearest neighbor, or a target locationQ, using the structure of the k-d tree, with respect to the Minkowski p-distance.query_ball_point(self, Q, r, p=2.0): This is a more sophisticated query that outputs all points within the Minkowski p-distancer, from a target locationQ.query_pairs(self, r, p=2.0): This finds all pairs of points whose Minkowski p-distance is, at most,r.query_ball_tree(self, other, r, p=2.0): This is similar toquery_pairs, but it finds all pairs of points from two different k-d trees, which are at a Minkowski p-distance of, at least,r.sparse_distance_matrix(self, other, max_distance): This computes a distance matrix between two kd-trees, leaving as zero, any distance greater thanmax_distance. The output is stored in a sparsedok_matrix.count_neighbors(self, other, r, p=2.0): This attribute is an implementation of the two-point correlation designed by Gray and Moore, to count the number of pairs of points from two different k-d trees, which are at a Minkowski p-distance not larger thanr. Unlikequery_ball, this attribute does not produce the actual pairs.
There is a faster implementation of this object created as an extension type in Cython, the cdef class cKDTree. The main difference is in the way the nodes are coded on each case:
- For
KDTree, the nodes are nested Python classes (the node being the top class, and both leafnode and innernode being subclasses that represent different kinds of nodes in the tree). - For
cKDTree, the nodes are C-type malloc'd structs, not classes. This makes the implementation much faster, at a price of having less control over a possible manipulation of the nodes.
Let's use this idea to solve a point location problem, and at the same time revisit the Voronoi diagram from Lake Superior:
In [17]: from scipy.spatial import cKDTree, Voronoi, voronoi_plot_2d In [18]: vor = Voronoi(superior['vertices']); ....: tree = cKDTree(superior['vertices'])
First, we query for the previous dataset, of 100 random locations, the seeds that are closest to each of them:
In [19]: tree.query(points) Out[19]: (array([ 0.38942726, 0.05020313, 0.06987993, 0.2150344 , 0.16101652, 0.08485664, 0.33217896, 0.07993277, 0.06298875, 0.07428273, 0.1817608 , 0.04084714, 0.0094284 , 0.03073465, 0.01236209, 0.02395969, 0.17561544, 0.16823951, 0.24555293, 0.01742335, 0.03765772, 0.20490015, 0.00496507]), array([ 3, 343, 311, 155, 370, 372, 144, 280, 197, 144, 251, 453, 42 233, 232, 371, 280, 311, 0, 307, 507, 49, 474, 370, 114, 5, 1, 372, 285, 150, 361, 84, 43, 98, 418, 482, 155, 144, 371, 113, 91, 3, 453, 91, 311, 412, 155, 156, 251, 251, 22, 179, 394, 189, 49, 405, 453, 506, 407, 36, 308, 33, 81, 46, 301, 144, 280, 409, 197, 407, 516]))
Note the output is a tuple with two numpy.array: the first one indicates the distances of each point to its closest seed (their nearest neighbors), and the second one indicates the index of the corresponding seed.
We can use this idea to represent the Voronoi diagram without a geometric description in terms of vertices, segments, and rays:
In [20]: X = np.linspace( 0.45, 0.50, 256); ....: Y = np.linspace(-0.40, -0.35, 256); ....: canvas = np.meshgrid(X, Y); ....: points = np.c_[canvas[0].ravel(), canvas[1].ravel()]; ....: queries = tree.query(points)[1].reshape(256, 256) In [21]: ax1 = plt.subplot(121, aspect='equal'); ....: voronoi_plot_2d(vor, ax=ax1); ....: plt.xlim( 0.45, 0.50); ....: plt.ylim(-0.40, -0.35) Out[21]: (-0.4, -0.35) In [22]: ax2 = plt.subplot(122, aspect='equal'); ....: plt.gray(); ....: plt.pcolor(X, Y, queries); ....: plt.plot(vor.points[:,0], vor.points[:,1], 'ro'); ....: plt.xlim( 0.45, 0.50); ....: plt.ylim(-0.40, -0.35); ....: plt.show()
This gives the following two different representations of the Voronoi diagram, in a small window somewhere along the North shore of Lake Superior:

A range searching problem tries to determine which objects of an input set intersect with a query object (that we call the
range). For example, when given a set of points in the plane, which ones are contained inside a circle of radius r centered at a target location Q? We can solve this sample problem easily with the attribute query_ball_point from a suitable implementation of a k-d tree. We can go even further if the range is an object formed by the union of a sequence of different balls. The same attribute gets the job done, as the following code illustrates:
In [23]: points = np.random.rand(320, 2); ....: range_points = np.random.rand(5, 2); ....: range_radii = 0.1 * np.random.rand(5) In [24]: tree = cKDTree(points); ....: result = set() In [25]: for k in range(5): ....: point = range_points[k] ....: radius = range_radii[k] ....: partial_query = tree.query_ball_point(point, radius) ....: result = result.union(set(partial_query)) ....: In [26]: print result set([130, 3, 166, 231, 40, 266, 2, 269, 120, 53, 24, 281, 26, 284]) In [27]: fig = plt.figure(); ....: plt.axes().set_aspect('equal') In [28]: for point in points: ....: plt.plot(point[0], point[1], 'ko') ....: In [29]: for k in range(5): ....: point = range_points[k] ....: radius = range_radii[k] ....: circle = plt.Circle(point, radius, fill=False) ....: fig.gca().add_artist(circle) ....: In [30]: plt.show()
This gives the following diagram, where the small dots represent the locations of the search space, and the circles are the range. The query is the set of points located inside of the circles, computed by our algorithm:

Problems in this setting vary from trivial to extremely complicated, depending on the input object types, range types, and query types. An excellent exposition of this subject is the survey paper Geometric Range Searching and its Relatives, published by Pankaj K. Agarwal and Jeff Erickson in 1999, by the American Mathematical Society Press, as part of the Advances in Discrete and Computational Geometry: proceedings of the 1996 AMS-IMS-SIAM joint summer research conference, Discrete and Computational geometry.
A dynamic problem is regarded as any of the problems in the previous two settings (static or query), but with the added challenge that objects are constantly being inserted or deleted. Besides solving the base problem, we need to take extra measures to assure that the implementation is efficient with respect to these changes.
To this effect, the implementations wrapped from the Qhull libraries in the module scipy.spatial are equipped to deal with the insertion of new points. We accomplish this by stating the option incremental=True, which basically suppresses the qhull control 'Qz', and prepares the output structure for these complex situations.
Let's illustrate this with a simple example. We start with the first ten vertices of Lake Superior, then we insert ten vertices at a time, and update the corresponding triangulation and Voronoi diagrams:
In [27]: from scipy.spatial import delaunay_plot_2d In [28]: small_superior = superior['vertices'][:9] In [29]: tri = Delaunay(small_superior, incremental=True); ....: vor = Voronoi(small_superior, incremental=True) In [30]: for k in range(4): ....: tri.add_points(superior['vertices'][10*(k+1):10*(k+2)-1]) ....: vor.add_points(superior['vertices'][10*(k+1):10*(k+2)-1]) ....: ax1 = plt.subplot(4, 2, 2*k+1, aspect='equal') ....: delaunay_plot_2d(tri, ax1) ....: ax1.set_xlim( 0.00, 1.00) ....: ax1.set_ylim(-0.70, -0.30) ....: ax2 = plt.subplot(4, 2, 2*k+2, aspect='equal') ....: voronoi_plot_2d(vor, ax2) ....: ax2.set_xlim(0.0, 1.0) ....: ax2.set_ylim(-0.70, -0.30) ....: In [4]: plt.show()
This displays the following diagram:

This field arose simultaneously among different groups of researchers seeking solutions to priori nonrelated problems. As it turns out, all the solutions they posed did actually have an important common denominator, they were obtained upon representing objects by means of parametric curves, parametric surfaces, or regions bounded by those. These scientists ended up unifying their techniques over the years, to finally define the field of numerical computational geometry. In this journey, the field received different names: machine geometry, geometric modeling, and the most widespread computer aided geometric design (CAGD).
It is used in computer vision, for example, for 3D reconstruction and movement outline. It is widely employed for the design and qualitative analysis of the bodies of automobiles, aircraft, or watercraft. There are many computer-aided design (CAD) software packages that facilitate interactive manipulation and solutions to many of the problems in this area. In this regard, any interaction with Python gets relegated to being part of the underlying computational engine behind the visualization or animation—which are none of the strengths of SciPy. For this reason, we will not cover visualization or animation applications in this book, and focus on the basic mathematics instead.
In that regard, the foundation of Numerical computational geometry is based on three key concepts: Bézier surfaces, Coons patches, and B-spline methods. In turn, the theory of Bézier curves plays a central role in the development of these concepts. They are the geometric standards for the representation of piecewise polynomial curves. In this section, we focus solely on the basic development of the theory of plane Bézier curves.
Tip
The rest of the material is also beyond the scope of SciPy, and we therefore leave its exposition to more technical books. The best source in that sense is, without a doubt, the book Curves and Surfaces for Computer Aided Geometric Design—A Practical Guide (5th ed.), by Gerald Farin, published by Academic Press under the Morgan Kauffman Series in Computer Graphics and Geometric Modeling.
It all starts with the de Casteljau algorithm to construct parametric equations of an arc of a polynomial of order 3. In the submodule matplotlib.path, we have an implementation of this algorithm using the class Path, which we can use to generate our own user-defined routines to generate and plot plane Bézier curves:
# file chapter6.py ...continued import matplotlib.pyplot as plt import matplotlib.patches as patches from matplotlib.path import Path def bezier_parabola(P1, P2, P3): return Path([P1, P2, P3], [Path.MOVETO, Path.CURVE3, Path.CURVE3]) def bezier_cubic(P1, P2, P3, P4): return Path([P1, P2, P3, P4], [Path.MOVETO, Path.CURVE4, Path.CURVE4, Path.CURVE4]) def plot_path(path, labels=None): Xs, Ys = zip(*path.vertices) fig = plt.figure() ax = fig.add_subplot(111, aspect='equal') ax.set_xlim(min(Xs)-0.2, max(Xs)+0.2) ax.set_ylim(min(Ys)-0.2, max(Ys)+0.2) patch = patches.PathPatch(path, facecolor='none', linewidth=2) ax.add_patch(patch) ax.plot(Xs, Ys, 'o--', color='blue', linewidth=1) if labels: for k in range(len(labels)): ax.text(path.vertices[k][0]-0.1, path.vertices[k][1]-0.1, labels[k]) plt.show()
Before we proceed, we need some explanation of the previous code:
- The de Casteljau algorithm for arcs of polynomials of order 2 is performed by creating a
Pathwith the three control points as vertices, and the list[Path.MOVETO, Path.CURVE3, Path.CURVE3]as code. This ensures that the resulting curve starts atP1in the direction given by the segmentP1P2, and ends atP3with direction given by the segmentP2P3. If the three points are collinear, we obtain a segment containing them all. Otherwise, we obtain an arc of parabola. - The de Casteljau algorithm for arcs of polynomials of order 3 is performed in a similar way to the previous case. We have four control points, and we create a
Pathwith those as vertices. The code is the list[Path.MOVETO, Path.CURVE4, Path.CURVE4, Path.CURVE4], which ensures that the arc starts atP1with direction given by the segmentP1P2. It also ensures that the arc ends atP4in the direction of the segmentP3P4.
Let's test it with a few basic examples:
In [1]: import numpy as np; ...: from chapter6 import * In [2]: P1 = (0.0, 0.0); ...: P2 = (1.0, 1.0); ...: P3 = (2.0, 0.0); ...: path_1 = bezier_parabola(P1, P2, P3); ...: plot_path(path_1, labels=['P1', 'P2', 'P3'])
This gives the requested arc of parabola:

In [3]: P4 = (2.0, -1.0); ...: P5 = (3.0, 0.0); ...: path_2 = bezier_cubic(P1, P2, P4, P5); ...: plot_path(path_2, labels=['P1', 'P2', 'P4', 'P5'])
This gives a nice arc of cubic as shown in the following diagram:

Higher degree curves are computationally expensive to evaluate. When complex paths are needed, we rather create them as a piecewise sequence of low order Bézier curves patched together—we call this object a Bézier spline. Notice that it is not hard to guarantee continuity on these splines. It is enough to make the end of each path the starting point of the next one. It is also easy to guarantee smoothness (at least up to the first derivative), by aligning the last two control points of one curve with the first two control points of the next one. Let's illustrate this with an example:
In [4]: Q1 = P5; ...: Q2 = (4.0, 0.0); ...: Q3 = (5.0, -1.0); ...: Q4 = (6.0, 0.0); ...: path_3 = bezier_cubic(P1, P2, P3, P5); ...: path_4 = bezier_cubic(Q1, Q2, Q3, Q4); ...: plot_path(Path.make_compound_path(path_3, path_4), labels=['P1','P2','P3','P5=Q1', 'P5=Q1','Q2','Q3', 'Q4'])
This gives the following Bézier spline:

A clear advantage of representing curves as Bézier splines arises when we need to apply an affine transformation to a curve. For instance, if we required a counter-clockwise rotated version of the last curve computed, instead of performing the operation over all points of the curve, we simply apply the transformation to the control points and repeat the de Casteljau algorithm on the new controls:
In [5]: def rotation(point, angle): ...: return (np.cos(angle)*point[0] - np.sin(angle)*point[1], ...: np.sin(angle)*point[0] + np.cos(angle)*point[1]) ...: In [6]: new_Ps = [rotation(P, np.pi/3) for P in path_3.vertices]; ...: new_Qs = [rotation(Q, np.pi/3) for Q in path_4.vertices]; ...: path_5 = bezier_cubic(*new_Ps); ...: path_6 = bezier_cubic(*new_Qs); ...: plot_path(Path.make_compound_path(path_5, path_6))
This displays the following result:

