Chapter 10. Low Rank Methods
In the last chapter, we lamented at the challenge of working with so many features. We saw that by letting each item be its own feature, we were able to express a lot of information about user preference and item affinity correlations, but we were in big trouble in terms of the curse of dimensionality. Combine this with the reality of very sparse features, and you’re in danger. Here we will instead turn to smaller feature spaces. By representing users and items as low dimensional vectors, we can capture the complex relationships between them in a more efficient and effective way. This allows us to generate more personalized and relevant recommendations for users, while also reducing the computational complexity of the recommendation process.
In this chapter, we will explore the use of low-dimensional embeddings in and discuss the benefits and some of the implementation details of this approach. We will also look at some code in JAX for how you can use modern gradient-based optimization to reduce the dimension of your item or user representations.
Latent Spaces
We are already familiar with feature spaces, which are usually categorical or vector-valued direct representations of the data. This can be the raw red, green, and blue values of an image, counts of items in a histogram, or attributes of an object like length, width, and height. Latent features on the other hand do not represent any specific real value feature of the items, but are initialized randomly and then learned to suit a task. The GloVe embeddings we discussed earlier were one such example of a latent vector that was learned to represent the log-count of words. Here we will cover more ways to generate these latent features or embeddings.
Focus on your Strangths
This chapter relies heavily on linear algebra, so knowledge about vectors, dot products, and norms of vectors are good to read up on before proceeding. Having an understanding of matrices, and rank of matrices are will also be useful.
Consider this reference by Gilbert Strang: best references.
One of the reasons latent spaces are so popular is that they are usually lower in dimension than the features they represent. For example, if the User-Item rating matrix or interaction matrix (where the matrix entries are 1 if a User has interacted with an item) is NxM dimensional, then factorizing the matrix into latent factors of NxK and KxM where K is much smaller then N or M is an approximation the missing entries – this is because we’re relaxing the factorization. K is being smaller than N or M is usually called an information bottleneck. That is, we are forcing the matrix to be made up of a much smaller matrix. This means the machine learning model has to make up missing entries, which might be good for recommender systems. As long as Users have interacted with enough similar items, by forcing the system to have a lot less capacity in terms of degrees of freedom, then it can completely reconstruct the matrix, and the missing entries tend to get filled by similar items.
Let’s see what happens, for example, when we factor a user item matrix of 4x4 into a 4x2 and a 2x4 vector using Singular Value Decomposition (SVD).
We are supplying a matrix whose rows are users and columns are items. For example, row 0 is [1, 0, 0, 1] which means user 0 has selected item 0 and item 3. These can be ratings or purchases. Now let’s look at some code:
importnumpyasnpa=np.array([[1,0,0,1],[1,0,0,0],[0,1,1,0],[0,1,0,0]])u,s,v=np.linalg.svd(a,full_matrices=False)# Set the last two eigenvalues to 0.s[2:4]=0(s)b=np.dot(u*s,v)(b)# These are the eigenvalues with the smallest two set to 0.s=[1.618033991.618033990.0.]# This is the newly reconstructed matrix.b=[[1.170820390.0.0.7236068][0.72360680.0.0.4472136][0.1.170820390.72360680.][0.0.72360680.44721360.]]
Notice that the user in row 1 now has a score for an item in column 3, and the user in row 3 now has a positive score in for the item column 2. This phenomenon is generally known as matrix completion, and is a good property for recommender systems to have because now we get to recommend new items to users. The general method of forcing the machine learning to go through a bottleneck that is smaller than the size of the matrix that it is trying to reconstruct is known as a low-rank approximation because the rank of the approximation is 2 but the rank of the original user-item matrix is 4.
What is the rank of a matrix?
An NxM matrix may be considered as N row vectors (corresponding to users) and M column vectors (corresponding to items). When you consider the N vectors of dimension M, the rank of the matrix is the volume of the polyhedron defined by the N vectors in M dimensions. This is often different than how we talk about rank of matrices, however. While it’s the most natural and precise definition, we instead say it’s the “minimum number of dimensions necessary to represent the vectors of the matrix”.
We will cover SVD in more detail later in the chapter, this was just to whet your appetite to how latent spaces are related to recommender systems.
Dot product similarity
In section 1 we introduced similarity measures, but now we return to the dot-product in the context of similarity because of their increased importance in latent-spaces – after all, latent-spaces are build on the assumption that distance is similarity.
The dot product similarity is meaningful in recommendation systems because it provides a geometric interpretation of the relationship between users and items in the latent space (or potentially items and items, users and users, etc.). In the context of recommendation systems, the dot product can be seen as a projection of one vector onto another, indicating the degree of similarity or alignment between the user’s preferences and the item’s characteristics.
To understand the geometric significance of the dot product, consider two vectors, u and p, representing the user and the product in the latent space, respectively. The dot product of these two vectors can be defined as:
where ||u|| and ||p|| represent the magnitudes of the user and product vectors, and θ is the angle between them. The dot product is thus a measure of the projection of one vector onto another, which is scaled by the magnitudes of both vectors.
The cosine similarity, which is another popular similarity measure in recommendation systems, is derived directly from the dot product:
The cosine similarity ranges from -1 to 1, where -1 indicates completely dissimilar preferences and characteristics. 0 indicates no similarity, and 1 indicates perfect alignment between the user’s preferences and the product’s characteristics. In the context of recommendation systems, cosine similarity provides a normalized measure of similarity that is invariant to the magnitudes of the user and product vectors. Note that the choice of using cosine similarity vs L2 distance depends on the type of embeddings you’re using, and how you optimize the computations. Often in practice the only important thing is the relative values.
The geometric interpretation of the dot product (and cosine similarity) in recommendation systems is that it captures the alignment between user preferences and product characteristics. If the angle between the user and product vectors is small, it means that the user’s preferences align well with the product’s characteristics, leading to a higher similarity score. Conversely, if the angle is large, the user’s preferences and product’s characteristics are dissimilar, resulting in a lower similarity score. By projecting user and item vectors onto each other, the dot product similarity can capture the degree of alignment between user preferences and item characteristics, allowing the recommendation system to identify items that are most likely to be relevant and appealing to the user.
Anecdotally, the dot product seems to capture popularity as very long vectors tend to be easy to project on anything that isn’t completely perpendicular or pointing away from them. As a result, there is a tradeoff between frequently recommending popular items with a large vector length from longer tail items that have smaller angular difference with cosine distance.

Figure 10-1. Cosine vs Dot product similarity
Figure 10-1 Considers two vectors, a and b. With Cosine similarity the vectors are unit length so the angle is just the measure of similarity. However, with dot product, a very long vector like c might be considered more similar to a than b even though the angle beween a and b is smaller due to the longer length of c. These long vectors tend to be very popular items that co-occur with many other items.
Co-occurrence models
In our Wikipedia co-occurrences examples, we determined that the co-occurrence structure between two items could be used to generate measures of similarity. We covered how pointwise mutual information can take the counts of co-occurrence, and make recommendations based on very high mutual information between an item in cart, and others.
When we discussed PMI, we found that it was not a distance metric, but still had some important similarity measures based on co-occurrence. Let’s return to this.
Recall from earlier that PMI is defined:
Now let’s consider the discrete co-occurrence distribution
where
There are a few different ways to measure distributional distance, each with different advantages. For our discussion we will not go deeply into the differences, and stick to the simplest but most appropriate. Hellinger distance is defined as:
The motivation of this process is that we now have a proper distance between items purely based on co-occurrences. We can use any dimension transformation or reduction on this geometry. Later we will see dimension reduction techniques that can utilize an arbitrary distance matrix and reduce the space to a lower dimensional embedding that approximates it.
What about measure spaces and information theory?
While we’re discussing distributions as we have, you may find yourself wondering: “is there a distance between distributions such that distributions are points in a latent space”. Oh, you weren’t wondering that? Well, ok. We’ll address it anyways.
The short answer is that there are a few ways to measure the differences between distributions. The most popular is Kullback–Leibler (KL) divergence, which is usually described in a Bayesian sense as the “amount of surprise in seeing the distribution P, when expecting the distribution Q”. However, KL is not a proper distance metric because it is asymmetric.
A symmetric distance metric, that has some other nice properties, is the Hellinger distance. Hellinger distance is effectively the 2-norm measure theoretic distance. Additionally, Hellinger distance naturally generalizes to discrete distributions.
In case you felt like this still didn’t scratch your itch for abstraction, we can also consider the Total Variation Distance, which is the limit in the space of Fisher Exact distance measures, which really means that it has all the nice properties of a distance of two distributions and no measure would ever consider them more dissimilar. Well, all the nice properties except for one – it’s not smooth. If you also want smoothness for differentiability you’ll need to approximate it via an offset.
If you ever need a distance between distributions, just use Hellinger.
Reducing the rank of a recommender problem
We’ve seen how, as the number of items and users grows, we very rapidly increase the dimensionality of our recommender problem. Because we’re representing each item and user as a column or vector, this scales like
Like many integers, many matrices can be factored into smaller matrices ; for integers smaller means of smaller value, for matrices smaller means of smaller dimensions. When we factor an
We seek a small value for
These methods are also very popular in other kinds of machine learning, but for our case, we’ll primarily be looking at factorizing ratings or interaction matrices.
There are frequently a few challenges to overcome when considering matrix factorization:
-
the matrix you wish to factor is sparse and often non-negative and/or binary.
-
the number of non-zero elements in each item vector can vary wildly like we saw in the Matthew effect.
-
factorizing matrices is cubic in complexity.
-
SVD and other full rank methods don’t work without imputation, which itself is complicated.
We’ll address these with some alternative optimization methods.
Optimizing for matrix factorization
The basic optimization we wish to execute is to approximate:
Notably, if you wish to optimize matrix entries directly, you’ll need to optimize
Alternating Least Squares
ALS seeks to switch back and forth between
where
-
each of these update rules require the gradient with respect to the relevant factor matrix
-
we update an entire factor matrix at a time, but we evaluate loss on the product of the factor matrices vs. the original matrix
-
we have a mysterious distance function
-
by the way we’ve constructed this optimization, we’re implicitly assuming that we’ll converge via this process to two well-approximating matrices (we often also impose a limit on number of iterations).
In JAX, these optimizations will be straightforward to implement, and we’ll see how similar the equational forms and the JAX code look.
Distance between matrices
There are a variety of ways to determine the distance between two matrices. As we’ve seen before, different distance measurements for vectors yield different interpretations from the underlying space. We won’t have as many complications for these computations but it’s worth a short observation. The most obvious approach is one we’ve already seen, observed mean squared error:
One useful alternative to the observed mean squared error is in the case when you have a single non-zero entry for a user-vector (alternatively a max rating). In that case, you could instead use a cross-entropy loss, which provides a logistic matrix factorization and thus probability estimate. For more details on how to implement this, you can read about it in this tutorial by Kyle Chung.
In our observed ratings, we expect (and see!) a large number of missing values and some item vectors with an over-represented number of ratings. This suggests we should consider non-uniformly weighted matrices. Next we’ll discuss how to account for this and other variants with regularization.
Regularlization for MF
WALS, or Weighted Alternating Least Squares, is similar to ALS, but it attempts to resolve these two data issues more gracefully. In WALS, the weight assigned to each observed rating is inversely proportional to the number of observed ratings for the user or item. This means that observed ratings for users or items with few ratings are given more weight in the optimization process.
We can apply these weights as a regularlization parameter in our eventual loss function,
Other regularization methods are important and popular as well for matrix factorization. Two powerful regularization techniques we’ll discuss are:
-
weight decay
-
gramian regularlization
As is often the case, weight decay is our
Similarly, matrix factorization has another regularlization technique which looks very standard, but in calculation, is quite different. This is via the gramians – essentially regularizing the size of the individual matrix entries, but there’s an elegant trick for the optimization. In particular, a gramian of a matrix
These regularlizations are the frobenius terms:
or expanded:
and the gramian terms
Finally, we have our loss function
Regularized Matrix Factorization Implementation
So far we’ve written a lot of math symbols, but all of those symbols have allowed us to arrive at a model which is extremely powerful. Regularized Matrix Factorization is an effective model for medium sized recommender problems. This model type is still in prod for many serious businesses. One classic issue with Matrix Factorization (MF) implementations is performance, but because we’re working in JAX which has extremely native GPU support, our implementation can actually be much more compact than what you may find in something like a pyTorch example.
Let’s work through how this model would look to predict ratings for a user-item matrix via this doubly regularized model with Gramians.
First we’ll do the simple setup. This will assume your ratings matrix is already on wandb:
importjaximportjax.numpyasjnpimportnumpyasnpimportpandasaspdimportos,json,wandb,mathfromjaximportgrad,jitfromjaximportrandomfromjax.experimentalimportsparsekey=random.PRNGKey(0)wandb.login()run=wandb.init(# Set entity to specify your username or team nameentity="wandb-un",# Set the project where this run will be loggedproject="jax-mf",# associate the runs to the right datasetconfig={"dataset":"MF-Dataset",})# note that we assume the dataset is a ratings table stored in wandbartifact=run.use_artifact('stored-dataset:latest')ratings_artifact=artifact.download()ratings_artifact_blob=json.load(open(os.path.join(ratings_artifact,'ratings.table.json')))ratings_artifact_blob.keys()# ['_type', 'column_types', 'columns', 'data', 'ncols', 'nrows']ratings=pd.DataFrame(# user_id, item_id, rating, unix_timestampdata=ratings_artifact_blob['data'],columns=ratings_artifact_blob['columns'])defstart_pipeline(df):returndf.copy()defcolumn_as_type(df,column:str,cast_type):df[column]=df[column].astype(cast_type)returndfdefrename_column_value(df,target_column,prior_val,post_val):df[target_column]=df[target_column].replace({prior_val:post_val})returndfdefsplit_dataframe(df,holdout_fraction=0.1):"""Splits a DataFrame into training and test sets.Args:df: a dataframe.holdout_fraction: fraction of dataframe rows to use in the test set.Returns:train: dataframe for trainingtest: dataframe for testing"""test=df.sample(frac=holdout_fraction,replace=False)train=df[~df.index.isin(test.index)]returntrain,testall_rat=(ratings.pipe(start_pipeline).pipe(column_as_type,column='user_id',cast_type=int).pipe(column_as_type,column='item_id',cast_type=int))defratings_to_sparse_array(ratings_df,user_dim,item_dim):indices=(np.array(ratings_df['user_id']),np.array(ratings_df['item_id']))values=jnp.array(ratings_df['rating'])return{'indices':indices,'values':values,'shape':[user_dim,item_dim]}defrandom_normal(pr_key,shape,mu=0,sigma=1,):return(mu+sigma*random.normal(pr_key,shape=shape))x=random_normal(pr_key=random.PRNGKey(1701),shape=(10000,),mu=1.0,sigma=3.0,)# these hyperparameters are pretty meaninglessdefsp_mse_loss(A,params):U,V=params['users'],params['items']rows,columns=A['indices']estimator=-(U@V.T)[(rows,columns)]square_err=jax.tree_map(lambdax:x**2,A['values']+estimator)returnjnp.mean(square_err)omse_loss=jit(sp_mse_loss)
Note that we’ve had to implement our own loss function here. This is a relatively straightforward MSE loss but it’s taking advantage of the sparse nature of our matrix. You may notice in the above that we’ve converted the matrix to a sparse representation, so it’s important that our loss function can not only take advantage of that representation, but that it’s written to utilize the JAX device arrays and mapping/jitting.
Is that loss function really right?
If you’re curious about this loss function which appeared like magic, I understand. During the writing of this book I was extremely uncertain what the best implementation of this loss function that leverages JAX would look like. There’s actually many reasonable approaches to this kind of optimization. To that end, I wrote a public experiment to benchmark several different approaches here.
Next, we need to build some model objects to handle our MF state as we train. This code, while essentially mostly template code, will set us up well to feed the model into a training loop in a relatively memory efficient way. This model was trained on 100M entries for a few thousand epochs on a Macbook Pro in less than a day.
classCFModel(object):"""Simple class that represents a collaborative filtering model"""def__init__(self,metrics:dict,embeddings:dict,ground_truth:dict,embeddings_parameters:dict,prng_key=None):"""Initializes a CFModel.Args:"""self._metrics=metricsself._embeddings=embeddingsself._ground_truth=ground_truthself._embeddings_parameters=embeddings_parametersifprng_keyisNone:prng_key=random.PRNGKey(0)self._prng_key=prng_key@propertydefembeddings(self):"""The embeddings dictionary."""returnself._embeddings@embeddings.setterdefembeddings(self,value):self._embeddings=value@propertydefmetrics(self):"""The metrics dictionary."""returnself._metrics@propertydefground_truth(self):"""The train/test dictionary."""returnself._ground_truthdefreset_embeddings(self):"""Clear out embeddings state."""prng_key1,prng_key2=random.split(self._prng_key,2)self._embeddings['users']=random_normal(prng_key1,[self._embeddings_parameters['user_dim'],self._embeddings_parameters['embedding_dim']],mu=0,sigma=self._embeddings_parameters['init_stddev'],)self._embeddings['items']=random_normal(prng_key2,[self._embeddings_parameters['item_dim'],self._embeddings_parameters['embedding_dim']],mu=0,sigma=self._embeddings_parameters['init_stddev'],)defmodel_constructor(ratings_df,user_dim,item_dim,embedding_dim=3,init_stddev=1.,holdout_fraction=0.2,prng_key=None,train_set=None,test_set=None,):ifprng_keyisNone:prng_key=random.PRNGKey(0)prng_key1,prng_key2=random.split(prng_key,2)if(train_setisNone)and(test_setisNone):train,test=(ratings_df.pipe(start_pipeline).pipe(split_dataframe,holdout_fraction=holdout_fraction))A_train=(train.pipe(start_pipeline).pipe(ratings_to_sparse_array,user_dim=user_dim,item_dim=item_dim))A_test=(test.pipe(start_pipeline).pipe(ratings_to_sparse_array,user_dim=user_dim,item_dim=item_dim))elif(train_setisNone)^(test_setisNone):raise('Must send train and test if sending one')else:A_train,A_test=train_set,test_setU=random_normal(prng_key1,[user_dim,embedding_dim],mu=0,sigma=init_stddev,)V=random_normal(prng_key2,[item_dim,embedding_dim],mu=0,sigma=init_stddev,)train_loss=omse_loss(A_train,{'users':U,'items':V})test_loss=omse_loss(A_test,{'users':U,'items':V})metrics={'train_error':train_loss,'test_error':test_loss}embeddings={'users':U,'items':V}ground_truth={"A_train":A_train,"A_test":A_test}returnCFModel(metrics=metrics,embeddings=embeddings,ground_truth=ground_truth,embeddings_parameters={'user_dim':user_dim,'item_dim':item_dim,'embedding_dim':embedding_dim,'init_stddev':init_stddev,},prng_key=prng_key,)mf_model=model_constructor(all_rat,user_count,item_count)
We should also set this up to log nicely to wandb so it’s easy to understand what is happening during training:
deftrain():run_config={# These will be hyperparameters we will tune via wandb'emb_dim':10,# Latent dimension'prior_std':0.1,# Std dev around 0 for weights initialization'alpha':1.0,# Learning rate'steps':1500,# Number of training steps}withwandb.init()asrun:run_config.update(run.config)model_object=model_constructor(ratings_df=all_rat,user_dim=user_count,item_dim=item_count,embedding_dim=run_config['emb_dim'],init_stddev=run_config['prior_std'],prng_key=random.PRNGKey(0),train_set=mf_model.ground_truth['A_train'],test_set=mf_model.ground_truth['A_test'])model_object.reset_embeddings()# Ensure we are starting from priorsalpha,steps=run_config['alpha'],run_config['steps'](run_config)grad_fn=jax.value_and_grad(omse_loss,1)foriinrange(steps):# We perform one gradient updateloss_val,grads=grad_fn(model_object.ground_truth['A_train'],model_object.embeddings)model_object.embeddings=jax.tree_multimap(lambdap,g:p-alpha*g,# Basic update rule; JAX handles broadcasting for usmodel_object.embeddings,grads)ifi%1000==0:# Most output in wandb; little bit of logging(f'Loss step{i}: ',loss_val)(f"""Test loss:{omse_loss(model_object.ground_truth['A_train'],model_object.embeddings)}""")wandb.log({"Train omse":loss_val,"Test omse":omse_loss(model_object.ground_truth['A_test'],model_object.embeddings)})
Note that this is using the tree_multimap to handle broadcasting our update rule, and we’re using the jitted loss from before in the omse_loss call. Also, we’re calling value_and_grad so we can log the loss to wandb as we go. This is a common trick you’ll see for efficiently doing both without a callback.
You can finish this off and start the training with a sweep:
sweep_config={"name":"mf-test-sweep","method":"random","parameters":{"steps":{"min":1000,"max":3000,},"alpha":{"min":0.6,"max":1.75},"emb_dim":{"min":3,"max":10},"prior_std":{"min":.5,"max":2.0},},"metric":{'name':'Test omse','goal':'minimize'}}sweep_id=wandb.sweep(sweep_config,project="jax-mf",entity="wandb-un")wandb.init()train()count=50wandb.agent(sweep_id,function=train,count=count)
In this case the hyperparameter optimization is over our hyperparameters like embedding dimension and the priors (randomized matrices). Up until now, we have trained some MF models on our ratings matrix. Let’s now add regularlization and cross-validation.
Translating the math equations above directly into code:
defell_two_regularlization_term(params,dimensions):U,V=params['users'],params['items']N,M=dimensions['users'],dimensions['items']user_sq=jnp.multiply(U,U)item_sq=jnp.multiply(V,V)return(jnp.sum(user_sq)/N+jnp.sum(item_sq)/M)l2_loss=jit(ell_two_regularlization_term)defgramian_regularization_term(params,dimensions):U,V=params['users'],params['items']N,M=dimensions['users'],dimensions['items']gr_user=U.T@Ugr_item=V.T@Vgr_square=jnp.multiply(gr_user,gr_item)return(jnp.sum(gr_square)/(N*M))gr_loss=jit(gramian_regularization_term)defregularized_omse(A,params,dimensions,hyperparams):lr,lg=hyperparams['ell_2'],hyperparams['gram']losses={'omse':sp_mse_loss(A,params),'l2_loss':l2_loss(params,dimensions),'gr_loss':gr_loss(params,dimensions),}losses.update({'total_loss':losses['omse']+lr*losses['l2_loss']+lg*losses['gr_loss']})returnlosses['total_loss'],lossesreg_loss_observed=jit(regularized_omse)
We won’t dive super deep into learning rate schedulers, but we will do a simple decay:
deflr_decay(step_num,base_learning_rate,decay_pct=0.5,period_length=100.0):returnbase_learning_rate*math.pow(decay_pct,math.floor((1+step_num)/period_length))
Our updated train function will incorporate our new regularlizations – which come with some hyperparameters – and a bit of additional logging setup. This code is making it easy to log our experiment as it trains, and configures the hyperparameters to work with regularlization.
deftrain_with_reg_loss():run_config={# These will be hyperparameters we will tune via wandb'emb_dim':None,'prior_std':None,'alpha':None,# Learning rate'steps':None,'ell_2':1,#l2 regularlization penalization weight'gram':1,#gramian regularlization penalization weight'decay_pct':0.5,'period_length':100.0}withwandb.init()asrun:run_config.update(run.config)model_object=model_constructor(ratings_df=all_rat,user_dim=942,item_dim=1681,embedding_dim=run_config['emb_dim'],init_stddev=run_config['prior_std'],prng_key=random.PRNGKey(0),train_set=mf_model.ground_truth['A_train'],test_set=mf_model.ground_truth['A_test'])model_object.reset_embeddings()# Ensure we start from priorsalpha,steps=run_config['alpha'],run_config['steps'](run_config)grad_fn=jax.value_and_grad(reg_loss_observed,1,has_aux=True)# Tell JAX to expect an aux dict as outputforiinrange(steps):(total_loss_val,loss_dict),grads=grad_fn(model_object.ground_truth['A_train'],model_object.embeddings,dimensions={'users':user_count,'items':item_count},hyperparams={'ell_2':run_config['ell_2'],'gram':run_config['gram']}# JAX carries our loss dict along for logging)model_object.embeddings=jax.tree_multimap(lambdap,g:p-lr_decay(i,alpha,run_config['decay_pct'],run_config['period_length'])*g,# update with decaymodel_object.embeddings,grads)ifi%1000==0:(f'Loss step{i}:')(loss_dict)(f"""Test loss:{omse_loss(model_object.ground_truth['A_test'],model_object.embeddings)}""")loss_dict.update(# wandb takes the entire loss dictionary{"Test omse":omse_loss(model_object.ground_truth['A_test'],model_object.embeddings),"learning_rate":lr_decay(i,alpha),})wandb.log(loss_dict)sweep_config={"name":"mf-HPO-with-reg","method":"random","parameters":{"steps":{"value":2000},"alpha":{"min":0.6,"max":2.25},"emb_dim":{"min":15,"max":80},"prior_std":{"min":.5,"max":2.0},"ell_2":{"min":.05,"max":0.5},"gram":{"min":.1,"max":.75},"decay_pct":{"min":.2,"max":.8},"period_length":{"min":50,"max":500}},"metric":{'name':'Test omse','goal':'minimize'}}sweep_id=wandb.sweep(sweep_config,project="jax-mf",entity="wandb-un")run_config={# These will be hyperparameters we will tune via wandb'emb_dim':10,# Latent dimension'prior_std':0.1,'alpha':1.0,# Learning rate'steps':1000,# Number of training steps'ell_2':1,#l2 regularlization penalization weight'gram':1,#gramian regularlization penalization weight'decay_pct':0.5,'period_length':100.0}train_with_reg_loss()
The last step is to do this in a way that gives us confidence in the models we’re seeing. Unfortunately, it can be a bit tricky to setup cross-validation for Matrix Factorization problems, so we’ll need to make a few modifications to our data structures.
defsparse_array_concatenate(sparse_array_iterable):return{'indices':tuple(map(jnp.concatenate,zip(*(x['indices']forxinsparse_array_iterable)))),'values':jnp.concatenate([x['values']forxinsparse_array_iterable]),}classjax_df_Kfold(object):"""Simple class that handles Kfoldsplitting of a matrix as a dataframe and stores as sparse jarrays"""def__init__(self,df:pd.DataFrame,user_dim:int,item_dim:int,k:int=5,prng_key=random.PRNGKey(0)):self._df=dfself._num_folds=kself._split_idxes=jnp.array_split(random.permutation(prng_key,df.index.to_numpy(),axis=0,independent=True),self._num_folds)self._fold_arrays=dict()forfold_indexinrange(self._num_folds):# let's create sparse jax arrays for each fold pieceself._fold_arrays[fold_index]=(self._df[self._df.index.isin(self._split_idxes[fold_index])].pipe(start_pipeline).pipe(ratings_to_sparse_array,user_dim=user_dim,item_dim=item_dim))defget_fold(self,fold_index:int):assert(self._num_folds>fold_index)test=self._fold_arrays[fold_index]train=sparse_array_concatenate([vfork,vinself._fold_arrays.items()ifk!=fold_index])returntrain,test
Each hyperparam setup should yield loss for each fold, so within the wandb.init, we build a model with each fold, thus:
forjinnum_folds:train,test=folder.get_fold(j)model_object_dict[j]=model_constructor(ratings_df=all_rat,user_dim=user_count,item_dim=item_count,embedding_dim=run_config['emb_dim'],init_stddev=run_config['prior_std'],prng_key=random.PRNGKey(0),train_set=train,test_set=test)
At each step, we’d like to not only compute the gradient for the training and evaluate on the test, we’d like to also compute gradients for all folds, eval on all the tests, and produce the relevant errors.
foriinrange(steps):loss_dict={"learning_rate":step_decay(i)}forj,Minmodel_object_dict.items():(total_loss_val,fold_loss_dict),grads=grad_fn(M.ground_truth['A_train'],M.embeddings,dimensions={'users':942,'items':1681},hyperparams={'ell_2':run_config['ell_2'],'gram':run_config['gram']})M.embeddings=jax.tree_multimap(lambdap,g:p-step_decay(i)*g,M.embeddings,grads)
Logging should be losses per fold, and the aggregate loss should be the target metric. This is because each fold is an independent optimization of the model parameters, however we wish to see aggregate behavior across the folds.
fold_loss_dict={f'{k}_fold-{j}':vfork,vinfold_loss_dict.items()}fold_loss_dict.update({f"Test omse_fold-{j}":omse_loss(M.ground_truth['A_test'],M.embeddings),})loss_dict.update(fold_loss_dict)loss_dict.update({"Test omse_mean":jnp.mean([vfork,vinloss_dict.items()ifk.startswith('Test omse_fold-')])})wandb.log(loss_dict)
Which we wrap up into one big training method:
deftrain_with_reg_loss_CV():run_config={# These will be hyperparameters we will tune via wandb'emb_dim':None,# Latent dimension'prior_std':None,# Standard deviation around 0 that our weights are initialized to'alpha':None,# Learning rate'steps':None,# Number of training steps'num_folds':None,# Number of CV Folds'ell_2':1,#hyperparameter for l2 regularlization penalization weight'gram':1,#hyperparameter for gramian regularlization penalization weight}withwandb.init()asrun:run_config.update(run.config)# This is how the wandb agent passes along paramsmodel_object_dict=dict()forjinrange(run_config['num_folds']):train,test=folder.get_fold(j)model_object_dict[j]=model_constructor(ratings_df=all_rat,user_dim=942,item_dim=1681,embedding_dim=run_config['emb_dim'],init_stddev=run_config['prior_std'],prng_key=random.PRNGKey(0),train_set=train,test_set=test)model_object_dict[j].reset_embeddings()# Ensure we are starting from priorsalpha,steps=run_config['alpha'],run_config['steps'](run_config)grad_fn=jax.value_and_grad(reg_loss_observed,1,has_aux=True)# Tell JAX to expect an aux dict as outputforiinrange(steps):loss_dict={"learning_rate":lr_decay(i,alpha,decay_pct=.75,period_length=250)}forj,Minmodel_object_dict.items():# Iterate through folds(total_loss_val,fold_loss_dict),grads=grad_fn(# compute gradients for one foldM.ground_truth['A_train'],M.embeddings,dimensions={'users':942,'items':1681},hyperparams={'ell_2':run_config['ell_2'],'gram':run_config['gram']})M.embeddings=jax.tree_multimap(# update weights for one foldlambdap,g:p-lr_decay(i,alpha,decay_pct=.75,period_length=250)*g,M.embeddings,grads)fold_loss_dict={f'{k}_fold-{j}':vfork,vinfold_loss_dict.items()}fold_loss_dict.update(# loss calculation within fold{f"Test omse_fold-{j}":omse_loss(M.ground_truth['A_test'],M.embeddings),})loss_dict.update(fold_loss_dict)loss_dict.update({# average loss over all folds"Test omse_mean":np.mean([vfork,vinloss_dict.items()ifk.startswith('Test omse_fold-')]),"test omse_max":np.max([vfork,vinloss_dict.items()ifk.startswith('Test omse_fold-')]),"test omse_min":np.min([vfork,vinloss_dict.items()ifk.startswith('Test omse_fold-')])})wandb.log(loss_dict)ifi%1000==0:# Most output in wandb; little bit of logging to stdout(f'Loss step{i}:')(loss_dict)
and our final sweeps config:
sweep_config={"name":"mf-HPO-CV","method":"random","parameters":{"steps":{"value":2000},"num_folds":{"value":5},"alpha":{"min":2.0,"max":3.0},"emb_dim":{"min":15,"max":70},"prior_std":{"min":.75,"max":1.0},"ell_2":{"min":.05,"max":0.5},"gram":{"min":.1,"max":.6},},"metric":{'name':'Test omse_mean','goal':'minimize'}}sweep_id=wandb.sweep(sweep_config,project="jax-mf",entity="wandb-un")wandb.agent(sweep_id,function=train_with_reg_loss_CV,count=count)
That may seem like a lot of setup, but we’ve really acheived a lot here. We’ve initialized the model to optimize the two matrix factors, while simultaneously keeping the matrix elements small, and keeping the gramians small also.
Which brings us to our lovely images.
Output from HPO MF
Let’s have a quick look at what what the prior work has produced. First, we see (Figure 10-3) that our primary loss function, OMSE, is rapidly decreasing. This is great, but we should take a deeper look.

Figure 10-3. The loss during training
Let’s also have a quick check that our regularization parameters (Figure 10-4) are converging, and we see that our l2 regularization could probably still decrease if we were to continue for more epochs.

Figure 10-4. Regularization Params
We’d like to see our cross-validation layed out by fold and corresponding loss (Figure 10-5). This kind of plot is called a parallel coordinates chart; it’s lines correspond to different runs which are in correspondance with different choices of parameters, and it’s vertical axes are different metrics. The far right heatmap axis corresponds to the overall total loss which we’re trying to minimize. In this case we alternate test loss on a fold and total loss on that fold. Lower numbers are better, and we hope to see individual lines consistent across their loss per fold (otherwise we may have a skewed dataset). We see that choices of hyper-parameters can interact with fold behavior, but in all of the low loss scenarios (at the bottom), we see a very high correlation between performance on different folds (the vertical axes in the plot).
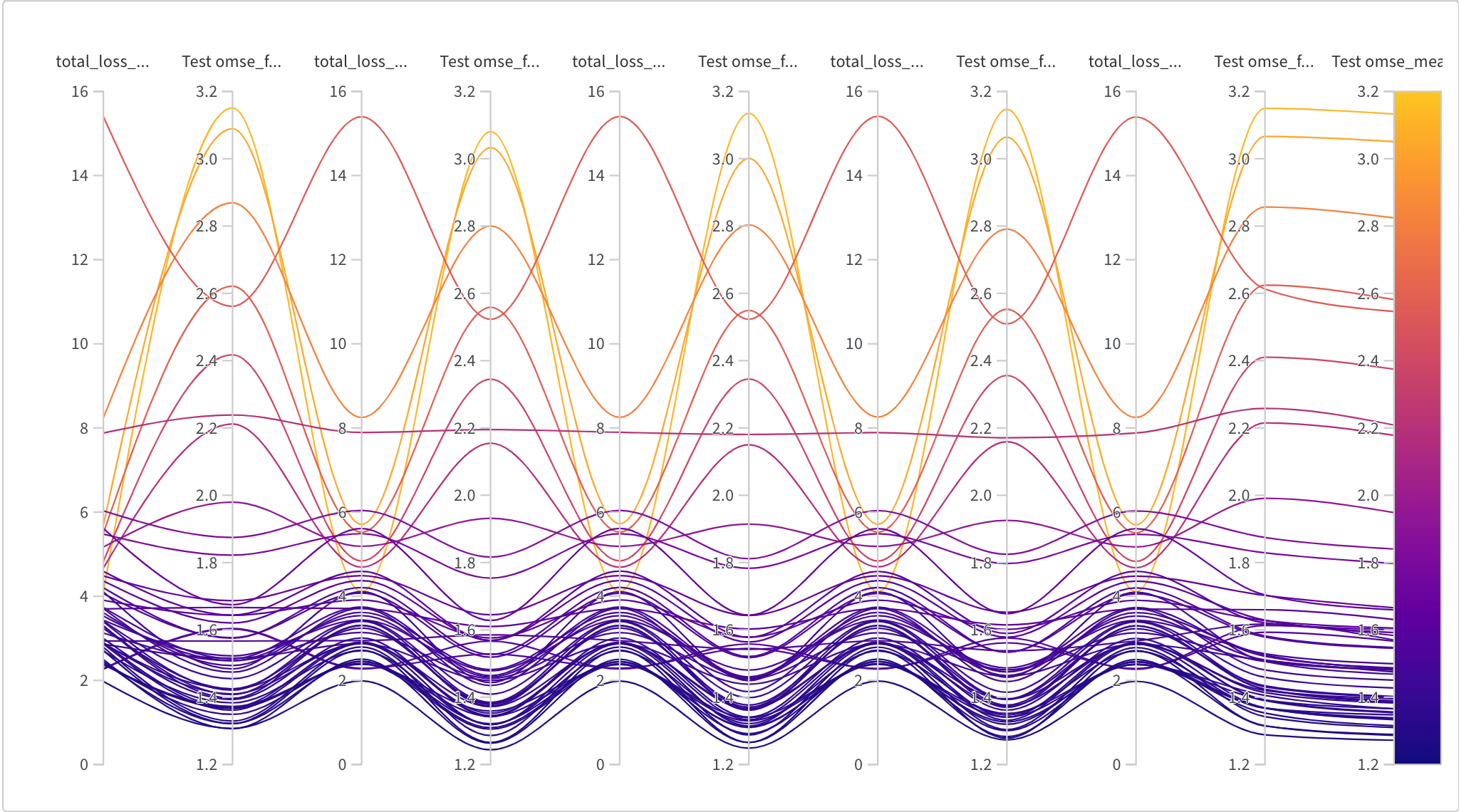
Figure 10-5. The loss during training
Next up, which choices of hyper-parameters have a strong effect on performance? In Figure 10-6 we have another parallel coordinates plot where the verticle axes correspond to different hyperparamaters. Generally we’re looking for which domains on the vertical axes correspond to low loss on the far right heatmap. We see that some of our hyperparameters like priors distribution and somewhat surprisingly ell_2 have virtually no effect. However, small embedding dimension and small gramian weight definitely do. A larger alpha also seems to correlate well with good performance.
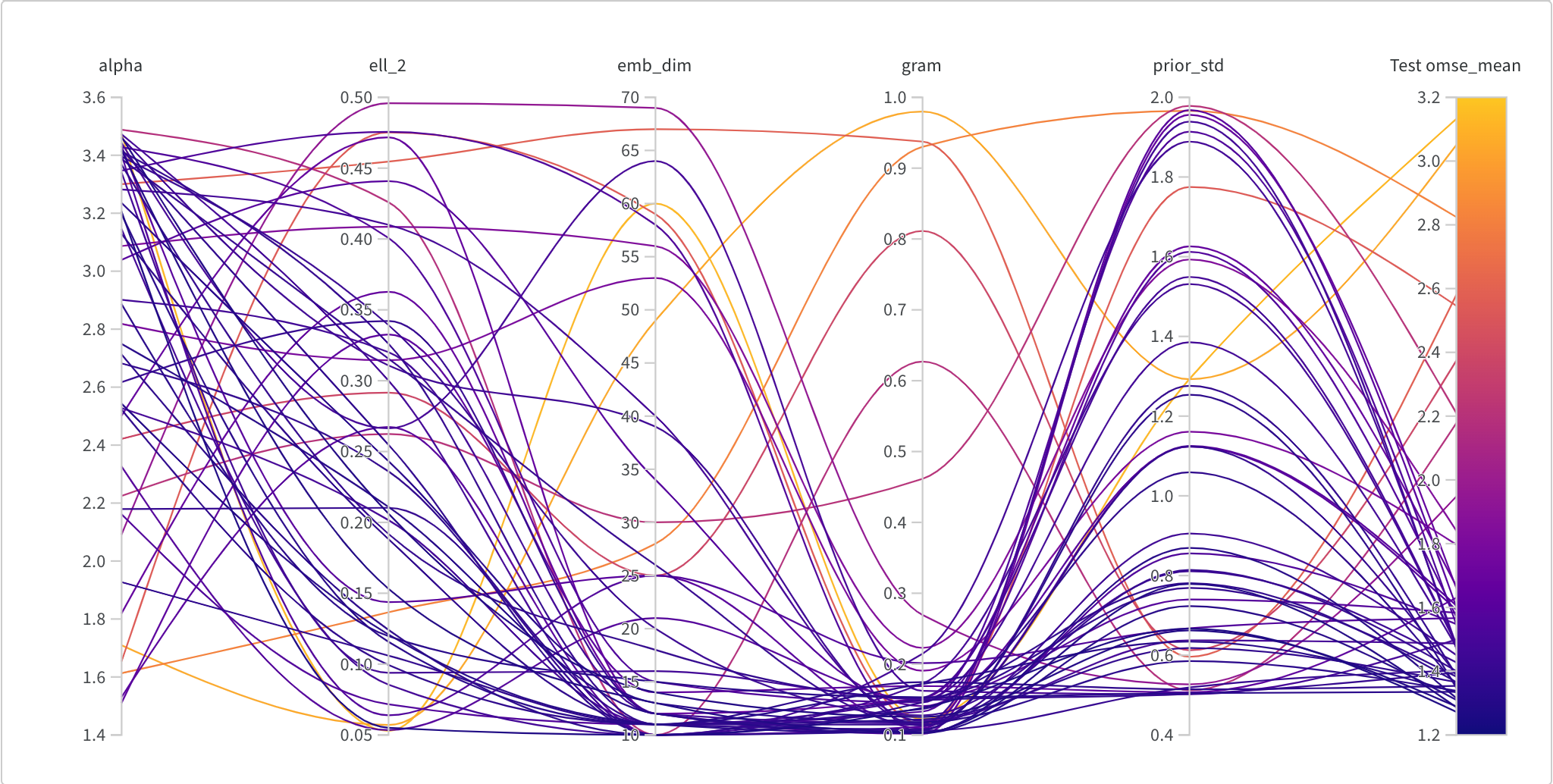
Figure 10-6. The loss during training
Finally, we see that as we do a Bayesian hyperparameter search, we really do improve our performance over time. The plot Figure 10-7, is a Pareto plot, where each dot in the scatter plot represents on run, and left to right is a time axis. The vertical axis is overall total loss, so lower is better, and it means that generally we’re converging towards better performance. The line inscribed along the bottom of the convex hull of the scatter points is the Pareto Frontier or “the best performance at that x value”. Since this is a time-series Pareto plot, this merely tracks the best performance in time. You may be wondering how and why we’re able to converge to better loss values in time. This is because we’ve conducted a Bayesian hyperparameter search, which means we select our hyperparameters from a independent Gaussians, and we update our priors for each parameter based on performance of previous runs; for an introduction to this method, this blog post covers it well. In a real setting, we’d see less monotonic behavior in this plot, but we’d be always hoping to improve.
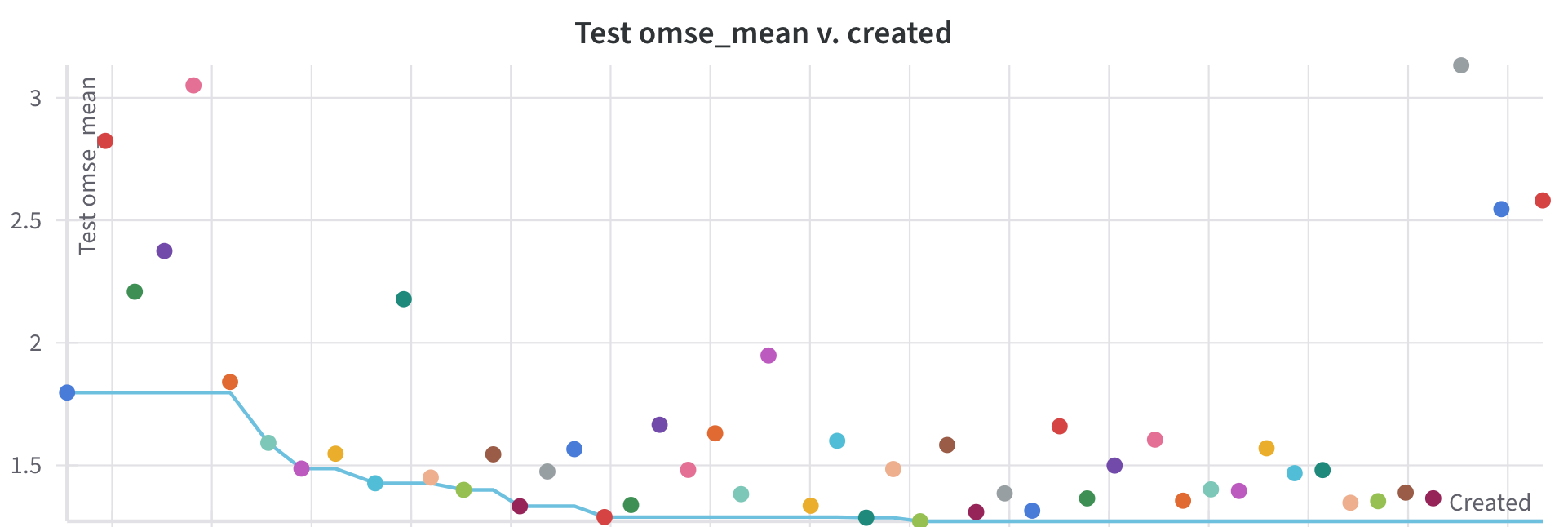
Figure 10-7. The loss during training
Pre-quential validation
If we were to put the above approach into practice, we would need to capture our trained models in a model registry for use in production.
Best practice is to establish a set of explicit evaluations to test a selection of models against. In your basic ML training, you’ve likely been encouraged to think about a validation dataset; these may take a lot of different forms, testing particular subsets of instances or features or even distributed across covariates in a known way.
One useful thing to do for recommendation systems, is to remember that they’re a fundamentally sequential dataset. With this in mind, let’s take another look at our ratings data. Later we will talk more about sequential recommenders, but while we’re talking about validation, it’s useful to mention how to take proper care.
Notice that all of our ratings have an associated timestamp. This means to build a proper validation set, it’s a good idea to take that from the end of our data.
However, you might be wondering: “when are different users active?” and “is it possible that the later timestamps are a biased selection of the ratings”. These are very important questions. To account this, we should do a holdout by user.
A simple way to generate this prequential dataset, a holdout-test dataset where the holdout is a contiguous series temporally after the holdout dataset, is to determine an approximate size you’d like for validation, say 10%, then group by user, then use rejection sampling with the rejection condition not largest timestamp.
Here’s a simple implementation for pandas using rejection sampling. This is not the most computationally efficient implementation, but it will get the job done:
defprequential_validation_set(df,holdout_perc=0.1):'''We utilize rejection sampling.Assign a probability to all observations, if they lie below thesample percentage AND they're the most recent still in the set, include.Otherwise return them and repeat.Each time, take no more than the remaining necessary to fill the count.'''count=int(len(df)*holdout_perc)sample=[]whilecount>0:df['p']=np.random.rand(len(df),1)#generate probabilitiesx=list(df.loc[~df.index.isin(sample)]# exclude already selected.sort_values(['unix_timestamp'],ascending=False)# sort by timestamp desc.groupby('user_id').head(1)# only allow the first in each group.query("p < @holdout_perc").index# grab the indices)rnd.shuffle(x)# make sure our previous sorting doesn't bias the users subsetsample+=x[:count]# add observations up to the remaining neededcount-=len(x[:count])# decrement the remaining neededdf.drop(columns=['p'],inplace=True)test=df.iloc[sample]train=df[~df.index.isin(test.index)]returntrain,test
This is an effective and important validation scheme for inherently sequential datasets.
WSABIE
Let’s focus back on optimizations and modifications one can do. Another optimization is to treat the Matrix Factorization problem as a single optimization.
The paper Wsabie: Scaling Up To Large Vocabulary Image Annotation also contains a factorization for just the item matrix. In this scheme, we replace the user matrix with a weighted sum of the items a user has affinity to. We will cover WSABIE and Warp loss in a later chapter on loss functions “WARP”. Representing a user as the average of items they like is a way of saving space and not needing a separate user matrix if there are large numbers of users.
Latent Space HPO
A completely alternative way to do HPO for RecSys is via the Latent Spaces themselves! Hyper-Parameter Optimization for Latent Spaces in Dynamic Recommender Systems attempts to modify the relative embeddings during each step to optimize the embedding model.
Dimension Reduction
Dimension reduction techniques are frequently employed in recommendation systems to decrease computational complexity and enhance the accuracy of recommendation algorithms. In this context, the primary concepts of dimension reduction for recommendation systems include:
Matrix factorization: The matrix factorization method decomposes the user-item interaction matrix
Singular value decomposition (SVD): SVD is a linear algebra technique that decomposes a matrix (A) into three matrices - the left singular vectors (U), the singular values (Σ), and the right singular vectors (V). SVD can be utilized for matrix factorization in recommendation systems, where the user-item interaction matrix is decomposed into a smaller number of latent factors. The mathematical representation of SVD is:
In practice though, rather than using a mathematical library to find the eigenvectors, folks might use the Power Iteration Method to discover the eigenvectors approximately. This method is far more scalable than a full dense SVD solve that is optimized for correctness and dense vectors.
importjaximportjax.numpyasjnpdefpower_iteration(a:jnp.ndarray)->jnp.ndarray:"""Returns an eigenvector of the matrix a.Args:a: a n x m matrix"""key=jax.random.PRNGKey(0)x=jax.random.normal(key,shape=(a.shape[1],1))foriinrange(100):x=a@xx=x/jnp.linalg.norm(x)returnx.Tkey=jax.random.PRNGKey(123)A=jax.random.normal(key,shape=[4,4])(A)[[0.528305530.3722206-1.2219944-0.10314374][1.47222220.47889313-1.29402981.0449569][0.237241850.3545859-0.172465-1.8011322][0.48642150.08039388-1.25408270.72071517]]S,_,_=jnp.linalg.svd(A)(S)[[-0.3757820.402698070.44086716-0.70870167][-0.7535970.0482972-0.655272840.01940039][0.20400880.91405433-0.157984940.31293103][-0.49925917-0.002500150.59270090.6320123]]x1=power_iteration(A)(x1.T)[[-0.35423845][-0.8332922][0.16189891][-0.39233655]]
Notice that the eigenvector returned by the power iteration is close to the first column of S but not quite. This is because the method is approximate. It relies on the fact that an eigenvector doesn’t change in direction when multiplied by the matrix. So by repeatedly multiplying by the matrix we eventually iterate onto an eigenvector. Also notice that we solved for column eigenvectors instead of the row eigenvectors. In this example, the columns are users and rows are items. It is important to play with transposed matrices as a lot of machine learning involves reshaping and transposing matrices, so getting used to them early is an important skill.
Eigenvector examples
Here’s a nice exercise for the reader: the second eigenvector is computed by subtracting out the first eigenvector after the matrix multiply. This is telling the algorithm to ignore any component along the first eigenvector in order to compute the second eigenvector. It would be a fun exercise to hop over to Colab and try computing the second eigenvector.
Extending this to sparse vector representations is another interesting exercise as it allows you to start computing the eigenvectors of sparse matrices which is usually the form of matrix that recommender systems use.
Next, we construct a recommendation for a user by creating a column and then taking the dot product with all the eigenvectors and finding the closest using the dot product. We then find all the highest scoring entries in the eigenvector that the user hasn’t seen and return them as recommendations. So in the example above, if the eigenvector x1 was the closest to the user column, then the best item to recommend would be item 3 because it is the largest component in the eigenvector and thus rated most highly if the user is closest to the eigenvector x_1. Here’s what this looks like in code:
importjaximportjax.numpyasjnpdefrecommend_items(eigenvectors:jnp.ndarray,user:jnp.ndarray)->jnp.ndarray:"""Returns an ordered list of recommend items for the user.Args:eigenvectors: a nxm eigenvector matrixuser: a user vector of size m."""score_eigenvectors=jnp.matmul(eigenvectors.T,user)which_eigenvector=jnp.argmax(score_eigenvectors)closest_eigenvector=eigenvectors.T[which_eigenvector]scores,items=jax.lax.top_k(closest_eigenvector,3)returnscores,itemsS=jnp.array([[-0.375782,0.40269807],[-0.753597,0.0482972],[0.2040088,0.91405433],[-0.49925917,-0.00250015]])u=jnp.array([-1,-1,0,0]).reshape(4,1)scores,items=recommend_items(S,u)(scores)[0.2040088-0.375782-0.49925917](items)[203]
In this example, we have a user who has downvoted item 0 and item 1. The closest column eigenvector is therefore column 0. We then select the closest eigenvector to the user and order the entries and recommend item 2 to the user which is the highest scoring entry that the user has not seen.
Two techniques that aim to extract the most relevant features from the user-item interaction matrix and reduce its dimensionality, which can improved performance.
-
Principal component analysis (PCA): PCA is a statistical technique that transforms the original high-dimensional data into a lower-dimensional representation while retaining the most important information. PCA can be applied to the user-item interaction matrix to reduce the number of dimensions and improve the computational efficiency of the recommendation algorithm.
-
Non-negative matrix factorization (NMF): NMF is a technique that decomposes the non-negative user-item interaction matrix
Matrix factorization techniques can be further extended to incorporate additional information, such as item content or user demographic data, through the use of side information. Side information can be employed to augment the user-item interaction matrix, allowing for more accurate and personalized recommendations.
Furthermore, matrix factorization models can be extended to handle implicit feedback data, where the absence of interaction data is not equivalent to the lack of interest. By incorporating additional regularization terms into the objective function, matrix factorization models can learn a more robust representation of the user-item interaction matrix, leading to better recommendations for implicit feedback scenarios.
Consider a recommendation system that employs matrix factorization to model the user-item interaction matrix. If the system comprises a large number of users and items, the resulting factor matrices can be high-dimensional and computationally expensive to process. However, by using dimension reduction techniques like SVD or PCA, the algorithm can reduce the dimensionality of the factor matrices while preserving the most important information about the user-item interactions. This enables the algorithm to generate more efficient and accurate recommendations, even for new users or items with limited interaction data.
Isometric embeddings
Isometric embeddings are a specific type of embedding that maintain distances between points in high-dimensional space when mapping them onto a lower-dimensional space. The term “isometric” signifies that the distances between points in the high-dimensional space are preserved precisely in the lower-dimensional space, up to a scaling factor.
In contrast to other types of embeddings, such as linear or non-linear embeddings, which may distort the distances between points, isometric embeddings are preferable in numerous applications where distance preservation is essential. For example, in machine learning, isometric embeddings can be employed to visualize high-dimensional data in two or three dimensions, while preserving the relative distances between the data points. In natural language processing, isometric embeddings can be utilized to represent the semantic similarities between words or documents, while maintaining their relative distances in the embedding space.
One popular technique for generating isometric embeddings is called Multi-dimensional Scaling (MDS). MDS operates by computing pairwise distances between the data points in the high-dimensional space and then determining a lower-dimensional embedding that preserves these distances. The optimization problem is generally formulated as a constrained optimization problem, where the objective is to minimize the difference between the pairwise distances in the high-dimensional space and the corresponding distances in the lower-dimensional embedding. Mathematically we write:
Here,
Another approach for generating isometric embeddings is through the use of kernel methods, such as Kernel PCA or Kernel MDS. Kernel methods work by implicitly mapping the data points into a higher-dimensional feature space, where the distances between the points are easier to compute. The isometric embedding is then calculated in the feature space, and the resulting embedding is mapped back to the original space.
Isometric embeddings have been employed in recommendation systems to represent the user-item interaction matrix in a lower-dimensional space where the distances between the items are preserved. By preserving the distances between items in the embedding space, the recommendation algorithm can better capture the underlying structure of the data and provide more accurate and diverse recommendations.
Isometric embeddings can also be employed to incorporate additional information into the recommendation algorithm, such as item content or user demographic data. By using isometric embeddings to represent the items and the additional information, the algorithm can capture the similarities between items based on both the user-item interaction data and the item content or user demographics, leading to more accurate and diverse recommendations.
Moreover, isometric embeddings can also be used to address the cold-start problem in recommendation systems. By using the isometric embeddings to represent the items, the algorithm can make recommendations for new items based on their similarities to the existing items in the embedding space, even in the absence of user interactions.
In summary, isometric embeddings are a valuable technique in recommendation systems for representing the user-item interaction matrix in a lower-dimensional space where the distances between the items are preserved. Isometric embeddings can be generated using matrix factorization techniques and can be employed to incorporate additional information, address the cold-start problem, and improve the accuracy and diversity of recommendations.
Non-linear locally metrizable embeddings
Non-linear locally metrizable embeddings are yet another method to represent the user-item interaction matrix in a lower-dimensional space where the local distances between nearby items are preserved. By preserving the local distances between items in the embedding space, the recommendation algorithm can better capture the local structure of the data and provide more accurate and diverse recommendations.
Mathematically, let
One popular approach to generating non-linear locally metrizable embeddings in recommendation systems is through the use of autoencoder neural networks. Autoencoders work by mapping the high-dimensional user-item interaction matrix onto a lower-dimensional space through an encoder network, and then reconstructing the matrix back in the high-dimensional space through a decoder network. The encoder and decoder networks are trained jointly to minimize the difference between the input data and the reconstructed data, with the objective of capturing the underlying structure of the data in the embedding space:
Here,
Another approach for generating non-linear locally metrizable embeddings in recommendation systems is through the use of t-SNE (t-Distributed Stochastic Neighbor Embedding). t-SNE works by modeling the pairwise similarities between the items in the high-dimensional space, and then finding a lower-dimensional embedding that preserves these similarities. A more popular approach in modern times is UMAP, which instead attempts to fit a minimal manifold that preserves density in local neighborhoods. UMAP is an essential technique for finding low-dimensional representations in complex and high dimensional latent spaces; find it’s documentation [here](https://umap-learn.readthedocs.io/en/latest/auto_examples/plot_mnist_example.html) The optimization problem is typically formulated as a cost function C that measures the difference between the pairwise similarities in the high-dimensional space and the corresponding similarities in the lower-dimensional embedding:
Here,
Non-linear locally metrizable embeddings can also be used to incorporate additional information into the recommendation algorithm, such as item content or user demographic data. By using non-linear locally metrizable embeddings to represent the items and the additional information, the algorithm can capture the similarities between items based on both the user-item interaction data and the item content or user demographics, leading to more accurate and diverse recommendations.
Moreover, non-linear locally metrizable embeddings can also be used to address the cold-start problem in recommendation systems. By using the non-linear locally metrizable embeddings to represent the items, the algorithm can make recommendations for new items based on their similarities to the existing items in the embedding space, even in the absence of user interactions.
In summary, non-linear locally metrizable embeddings are a useful technique in recommendation systems for representing the user-item interaction matrix in a lower-dimensional space where the local distances between nearby items are preserved. Non-linear locally metrizable embeddings can be generated using techniques such as autoencoder neural networks or t-SNE, and can be used to incorporate additional information, address the cold-start problem, and improve the accuracy and diversity of recommendations.
Centered Kernel Alignment
When training neural networks, the latent space representations at each layer are expected to express correlation structures between the incoming signals. Frequently, these interstitial representations comprise a sequence of states transitioning from the initial layer to the final layer. One may naturally wonder ‘how do these representations change throughout the layers of the network’ and ‘how similar are these layers?”. Interestingly, for some architectures this question may yield deep insight into the network’s behavior.
This process of comparing layer representations is called correlation analysis and for an MLP with layers
Affinity and p-sale
As we’ve seen, Matrix factorization is a powerful dimension reduction technique that can yield an estimator for the probability of a sale (often shorted to p-sale). In matrix factorization, the goal has been to decompose this the historical data on user behavior and product sales matrix into two lower-dimensional matrices: one that represents user preferences and another that represents product characteristics. Now, let’s convert this MF model into an sale estimator.
Let
The probability of a sale, or equivalently a read, watch, eat, or click, can be predicted using matrix factorization by first decomposing the historical data matrix into user and product matrices, and then calculating a score that represents the likelihood of a user purchasing a given product. This score can be calculated using the dot product of the corresponding row in the user matrix and column in the product matrix, followed by a logistic function to transform the dot product into a probability score.
Mathematically, the probability of a sale for a user u and a product p can be represented as:
where sigmoid is the logistic function that maps the dot product of the user and product vectors to a probability score between 0 and 1:
and
The user and product matrices can be trained on the historical data using various matrix factorization algorithms, such as singular value decomposition (SVD), non-negative matrix factorization (NMF), or alternating least squares (ALS). Once the matrices are trained, the dot product and logistic function can be applied to new user-product pairs to predict the probability of a sale. The predicted probabilities can then be used to rank and recommend products to the user.
It’s worth highlight that, since the loss function for ALS is convex (meaning there is a single global minimum) when one fixes either the user or item matrix, the convergence can be very fast. In this method, the user matrix is fixed and the item matrix is solved for. Then the item matrix is fixed and the user matrix is solved for. The method alternates between the two solves and because the loss is convex in this regime, the method converges very quickly.
The dot product of the corresponding row in the user matrix and column in the product matrix represents the affinity score between the user and the product, or how well the user’s preferences match the product’s characteristics. However, this score alone may not be a sufficient predictor of whether the user will actually purchase the product.
The logistic function applied to the dot product in the matrix factorization model transforms the affinity score into a probability score, which represents the likelihood of a sale. This transformation takes into account additional factors beyond just the user’s preferences and the product’s characteristics, such as the overall popularity of the product, the user’s purchasing behavior, and any other relevant external factors. By incorporating these additional factors, matrix factorization is able to better predict the probability of a sale, rather than just an affinity score.
A comparison library (however not in JAX) for computing latent embeddings linearly is Factorization Machines. The formulation for a Factorization Machine is very similar to a GloVe embedding in that it also models the interaction between two vectors but the dot product can be used for regression or binary classification tasks. The method can also be extended to recommend more than two kinds of items beyond user and item.
In summary, matrix factorization produces probabilities of sale instead of just affinity scores by incorporating additional factors beyond just the user’s preferences and the product’s characteristics, and transforming the affinity score into a probability score using a logistic function.
Propensity Weighting for Recommendation System Evaluation
As we’ve seen, recommendation systems are evaluated based on user feedback, which is collected from the deployed recommendation system. However, this data is causally influenced by the deployed system, creating a feedback loop that may bias the evaluation of new models. This feedback loop can lead to confounding variables, making it difficult to distinguish between user preferences and the influence of the deployed system. If this surprises you, let’s consider for a moment what would have to be true for a recommendation system to not causally influence the actions users take and/or the outcomes that result from those actions – that would require things like “the recommendations are completely ignored by the user” and “the system makes recommendations at random”. Propensity weighting can mitigate some of the worst effects of this problem.
The performance of a recommender system depends on many factors, including user-item characteristics, contextual information, and trends, which can affect the quality of the recommendations and the user engagement. However, the influence can be mutual: the user interactions influence the recommender and vice-versa. Evaluating the causal effect of a recommender system on user behavior and satisfaction is therefore a challenging task, as it requires controlling for potential confounding factors, ie things that may affect both the treatment assignment (i.e., the recommendation strategy) and the outcome of interest (i.e., the user’s response to the recommendations).
Causal inference provides a framework for addressing these challenges. In the context of recommender systems, causal inference can help answer questions such as:
-
How does the choice of recommendation strategy affect user engagement, such as click-through rates, purchase rates, and satisfaction ratings?
-
What is the optimal recommendation strategy for a given user segment, item category, or context?
-
What are the long-term effects of a recommendation strategy on user retention, loyalty, and lifetime value?
We’ll round out this chapter by introducing one aspect of causal inference important to recommender systems, based on the concept of propensity score.
Propensity
In many data science problems, we are forced to contend with confounders and notably the correlation between those confounders and a target outcome. Depending on the setting, the confounder may be of a variety of forms. Interestingly in Recommendation Systems that confounder can be the RecSys itself! Offline evaluation of recommendation systems is subject to confounders derived from the item selection behavior of users and the deployed recommendation system. If this seems a bit circular, it kinda is. This is sometimes called closed-loop feedback. One approach to mitigation is propensity weighting, which aims to address this problem by considering each feedback in the corresponding stratum based on the estimated propensities. You may recall that propensity refers to the likelihood of a user seeing an item; by inversely weighting by this we can offset the selection bias. Compared to the standard offline holdout evaluation, this method attempts to represent the actual utility of the examined recommendation models.
Note
One other approach to mitigating selection bias that we won’t dive into is counterfactual evaluation which estimates the actual utility of a recommendation model with propensity-weighting techniques more similar to off-policy evaluation approaches in reinforcement learning. However, it often relies on accurate logging propensities in an open-loop setting where some random items are exposed to the user, which is not practical for most recommendation problems. If you have the option to include randomized recommendations to users for rating, this can help de-bias as well. One such setting where these methods may be combined is in RL-based recommenders that use explore-exploit methods like a multi-armed bandit or other structured randomization.
Inverse Propensity Scoring (IPS) is a propensity-based evaluation method that leverages importance sampling to account for the fact that the feedback collected from the deployed recommendation system is not uniformly random. The propensity score is a balancing factor that adjusts the observed feedback distribution conditioned on the propensity score. The IPS evaluation method is theoretically unbiased if open loop feedback can be sampled from all possible items uniformly at random. At the start of the book, we discussed the Matthew effect, or “the rich get richer” for recommendation systems; IPS is one way to combat this effect. Note the relationship here between the two ideas of the Matthew effect and Simpson’s paradox, when within different strata selection effects create significant biasing.
Propensity weighting is based on the idea that the probability of an item being exposed to a user by the deployed recommendation system (the propensity score) affects the feedback that is collected from that user. By reweighting the feedback based on the propensity scores, we can adjust for the bias introduced by the deployed system and obtain a more accurate evaluation of the new recommendation model.
To apply IPS, we need to estimate the propensity scores for each item-user interaction in the collected feedback dataset. This can be done by modeling the probability that the deployed system would have exposed the item to the user at the time of the interaction. One simple approach is to use the popularity of the item as a proxy for its propensity score. However, more sophisticated methods can be used to model the propensity scores based on user and item features, as well as the context of the interaction.
Once the propensity scores are estimated, we can reweight the feedback using importance sampling. Specifically, each feedback is weighted by the inverse of its propensity score, so that items that are more likely to be exposed by the deployed system are downweighted, while items that are less likely to be exposed are upweighted. This reweighting process approximates a counterfactual distribution of feedback expected from surfacing recommendations from a uniform distribution of popularity.
Finally, we can use the reweighted feedback to evaluate the new recommendation model using standard metrics for evaluation as we’ve seen in this chapter. The effectiveness of the new model is then compared to that of the deployed system using the reweighted feedback, providing a fairer and more accurate evaluation of the new model’s performance.
Simpson’s and mitigating confounding
Simpson’s paradox is predicated on the idea of a confounding variable which establishes strata within which we see (potentially misleading) covariation. This paradox arises when the association between two variables is investigated, but these variables are strongly influenced by a confounding variable. In the case of recommendation systems, this confounding variable is the deployed model’s characteristics and tendencies of selection. The propensity score is introduced as a measure of a system’s deviation from an unbiased open loop exposure scenario. It allows for the design and analysis of offline evaluation of recommendation models based on the observed closed loop feedback, mimicking some of the particular characteristics of the open loop scenario.
Traditional descriptions of Simpson’s paradox often suggest stratification – a well-known approach to identify and estimate causal effects by first identifying the underlying strata before investigating causal effects in each stratum. This approach enables the measurement of the potential outcome irrespective of the confounding variable. In the case of recommendation systems, this involves stratifying the observed outcome based on the possible values of the confounding variable, which is the deployed model’s characteristics.
The user-independent propensity score is estimated using a two-step generative process, where the prior probability that an item is recommended by the deployed model and the conditional probability that the user interacts with the item given that it is recommended are used. Based on a set of mild assumptions (but too mathematically technical to cover here), the user-independent propensity score can be estimated using maximum likelihood for each dataset.
We need to define the user-propensity score
where
With these estimates for propensity, we can then apply a simple inverse weighting
Doubly-robust estimation
DRE is a method combines two models: one that models the probability of receiving the treatment (i.e., being recommended an item by the deployed model), and one that models the outcome of interest (i.e., the user’s feedback on the item). The weights used in DR estimation depend on the predicted probabilities from both models. This method has the advantage that it can still provide unbiased estimates even if one of the models is misspecified.
The structural equations for a doubly robust estimator with propensity score weighting and outcome model are:
where
For a great introduction to these considerations, check out the article Give me a robust estimator - and make it a double!.
Summary
What a whirlwind! Latent spaces are one of the most important aspects of recommendation systems. They are the representations that we use to encode our users and items. Ultimately, latent spaces are about more than dimension reduction – they are about understanding a geometry in which measures of distance encode the meaning relevant to your ML task.
The world of embeddings and encoders runs deep. We’ve not had time to discuss CLIP embeddings (image + text), or the Poincare disk (naturally heirarchical distance measures). We didn’t dive deep into UMAP (a nonlinear density aware dimension reduction technique) or HNSW (a method for retrieval in latent spaces that respects local geometry well). Instead we point you at the (contemporaneously published) article by Vicki Boykis on embeddings, the essay and guide to constructing embeddings by Karel Minařík, or the beautiful visual guide to text embeddings by Meor Amer from Cohere.
We’re now equipped with representations, but next we need to optimize. We’re building personalized recommendation systems, so let’s define the metrics which measure our performance on our task.
