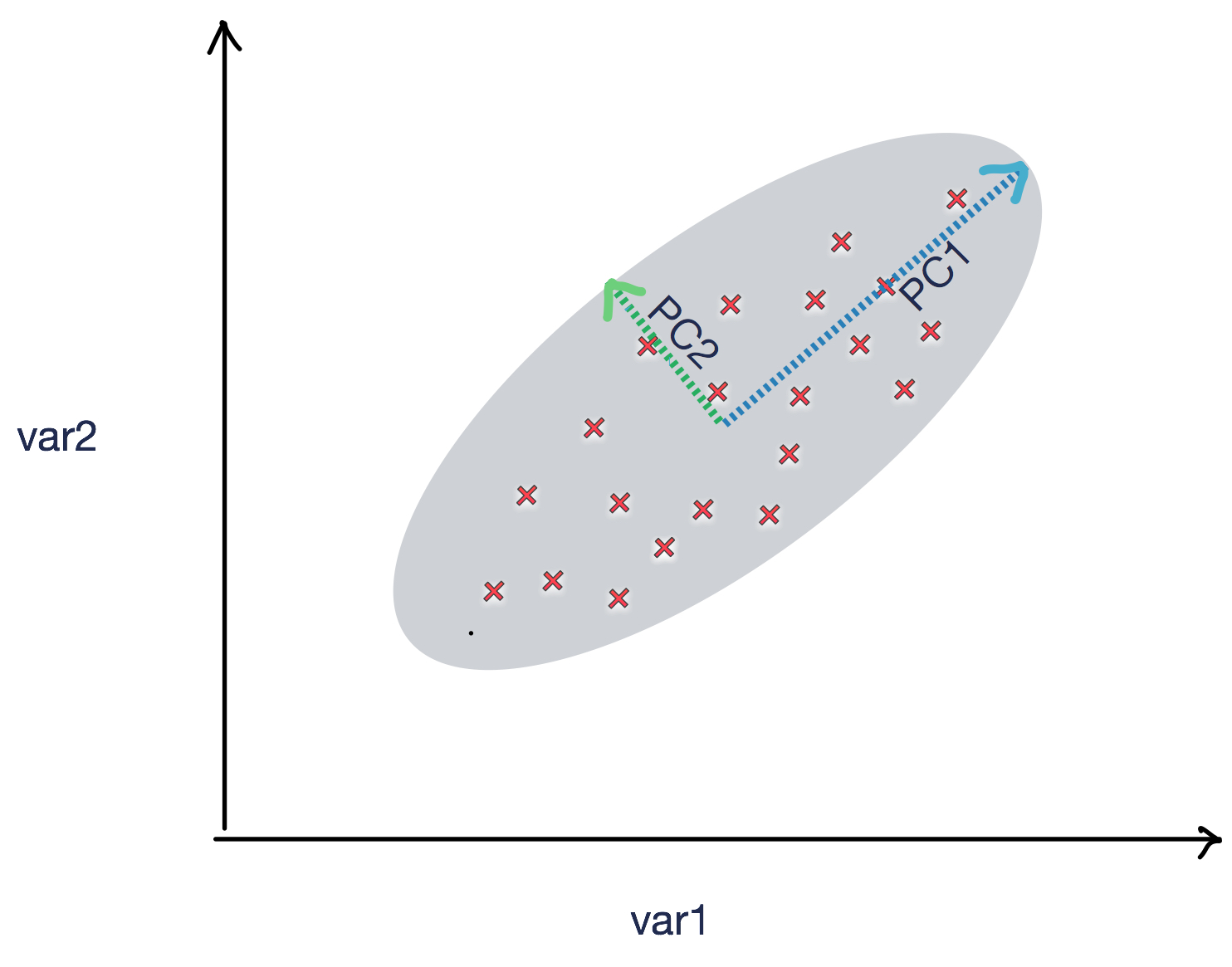
PCA is an unsupervised machine learning technique that can be used to reduce dimensions using linear transformation. In the following figure, we can see two principle components, PC1 and PC2, which show the shape of the spread of the data points. PC1 and PC2 can be used to summarize the data points with appropriate coefficients:
Let's consider the following code:
from sklearn.decomposition import PCA
iris = pd.read_csv('iris.csv')
X = iris.drop('Species', axis=1)
pca = PCA(n_components=4)
pca.fit(X)
Now let's print the coefficients of our PCA model:

Note that the original DataFrame has four features, Sepal.Length, Sepal.Width, Petal.Length, and Petal.Width. The preceding DataFrame specifies the coefficients of the four principal components, PC1, PC2, PC3, and PC4—for example, the first row specifies the coefficients of PC1 that can be used to replace the original four variables.
Based on these coefficients, we can calculate the PCA components for our input DataFrame X:
pca_df=(pd.DataFrame(pca.components_,columns=X.columns))
# Let us calculate PC1 using coefficients that are generated
X['PC1'] = X['Sepal.Length']* pca_df['Sepal.Length'][0] + X['Sepal.Width']* pca_df['Sepal.Width'][0]+ X['Petal.Length']* pca_df['Petal.Length'][0]+X['Petal.Width']* pca_df['Petal.Width'][0]
# Let us calculate PC2
X['PC2'] = X['Sepal.Length']* pca_df['Sepal.Length'][1] + X['Sepal.Width']* pca_df['Sepal.Width'][1]+ X['Petal.Length']* pca_df['Petal.Length'][1]+X['Petal.Width']* pca_df['Petal.Width'][1]
#Let us calculate PC3
X['PC3'] = X['Sepal.Length']* pca_df['Sepal.Length'][2] + X['Sepal.Width']* pca_df['Sepal.Width'][2]+ X['Petal.Length']* pca_df['Petal.Length'][2]+X['Petal.Width']* pca_df['Petal.Width'][2]
# Let us calculate PC4
X['PC4'] = X['Sepal.Length']* pca_df['Sepal.Length'][3] + X['Sepal.Width']* pca_df['Sepal.Width'][3]+ X['Petal.Length']* pca_df['Petal.Length'][3]+X['Petal.Width']* pca_df['Petal.Width'][3]
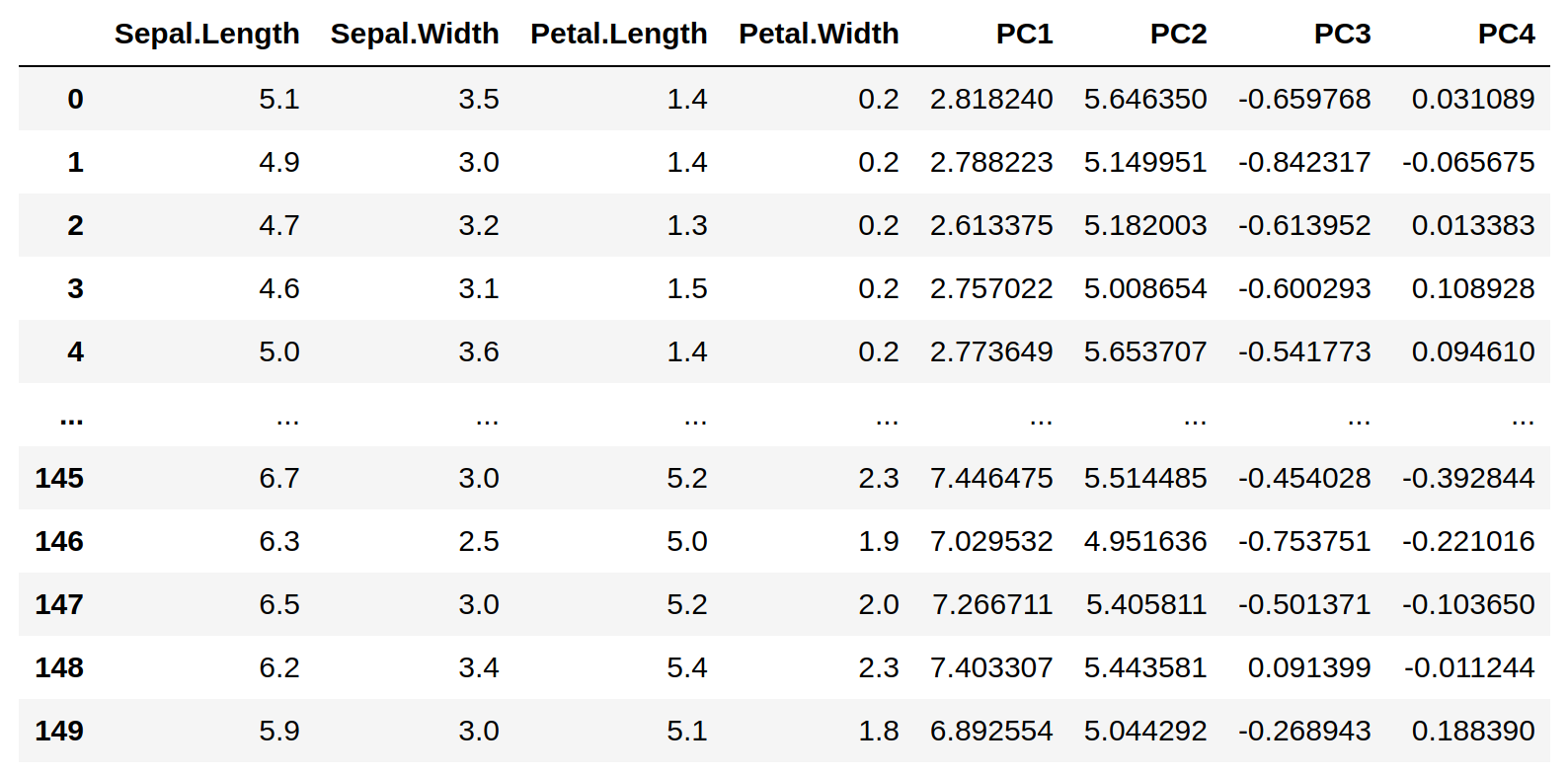
Now let's print X after the calculation of the PCA components:
Now let's print the variance ratio and try to understand the implications of using PCA:

The variance ratio indicates the following:
- If we choose to replace the original four features with PC1, then we will be able to capture about 92.3% of the variance of the original variables. We will introduce some approximations by not capturing 100% of the variance of the original four features.
- If we choose to replace the original four features with PC1 and PC2, then we will capture an additional 5.3 % of the variance of the original variables.
- If we choose to replace the original four features with PC1, PC2, and PC3, then we will now capture a further 0.017 % of the variance of the original variables.
- If we choose to replace the original four features with four principal components, then we will capture 100% of the variance of the original variables (92.4 + 0.053 + 0.017 + 0.005), but replacing four original features with four principal components is meaningless as we did not reduce the dimensions at all and achieved nothing.
