CHAPTER 2
Retrieval of Protein Sequence from UniProtKB
CS Mukhopadhyay and RK Choudhary
School of Animal Biotechnology, GADVASU, Ludhiana
2.1 INTRODUCTION
The Universal Protein Resource (UniProt) is a database of protein sequence and function, created by combining the Protein Information Resource‐Protein Sequence Database (PIR‐PSD), Swiss‐Prot, and TrEMBL databases. UniProt (www.uniprot.org/) has two sections: the Swiss‐Prot knowledgebase (it harbors fully annotated records) and the TrEMBL protein database (contains computationally analyzed records on proteins).
2.1.1 Features of UniProtKB/Swiss‐Prot
- Non‐redundancy of records.
- High level of integration of data deposited in different related databases (NCBI‐GenBank, EMBL, DDBJ for translated coding sequences).
- High level of manual curation.
- Contains more than 0.25 million entries.
2.1.2 Features of UniProtKB/TrEMBL
- Translations of nucleotide coding sequence (cds) in EMBL/NCBI‐GenBank/DDBJ.
- Automatic annotation.
- Contains more than 3.3 million entries.
2.2 OBJECTIVE
To download the amino acid sequence of protein (say, taurine sex‐determining region, Y‐encoded (SRY) peptide).
2.3 PROCEDURE
- Open the Expert Protein Analysis System (ExPASy) homepage: http://www.expasy.org/
- Locate the browser on the drop‐down menu “Query all databases” at the upper center portion of the page, and click on “Proteomics” (to obtain information from all relevant databases, such as Prosite, String, ENZYME, UniProtKB etc), or else select UniProtKb (Figure 2.1 below).
- Write the name of the protein and the species: “SRY Bos taurus” in the blank text box just beside “Find resources”.
- Click on “Search”.
- A list of search results is obtained in a table. Select the specific result: here it is “SRY_Bovin.”
- Click on the Entry (here “Q03255”) to get the detail of the sequence (see Figure 2.2).
- The newly opened page shows detailed information on the target protein, including names and origin, protein attributes, general annotation (i.e., comments), ontologies, sequence annotation (features), sequences, references, and so on.
- Click on FASTA to obtain the sequence in FASTA format.
- Select the sequence in FASTA format and copy and paste in a text file (see Figure 2.3).

FIGURE 2.1 Homepage of ExPASy server: select the “proteomics” option from the drop‐down menu for databases, and enter your protein name along with other keywords to begin search.

FIGURE 2.2 Click on the specific entry to open it in a separate window.
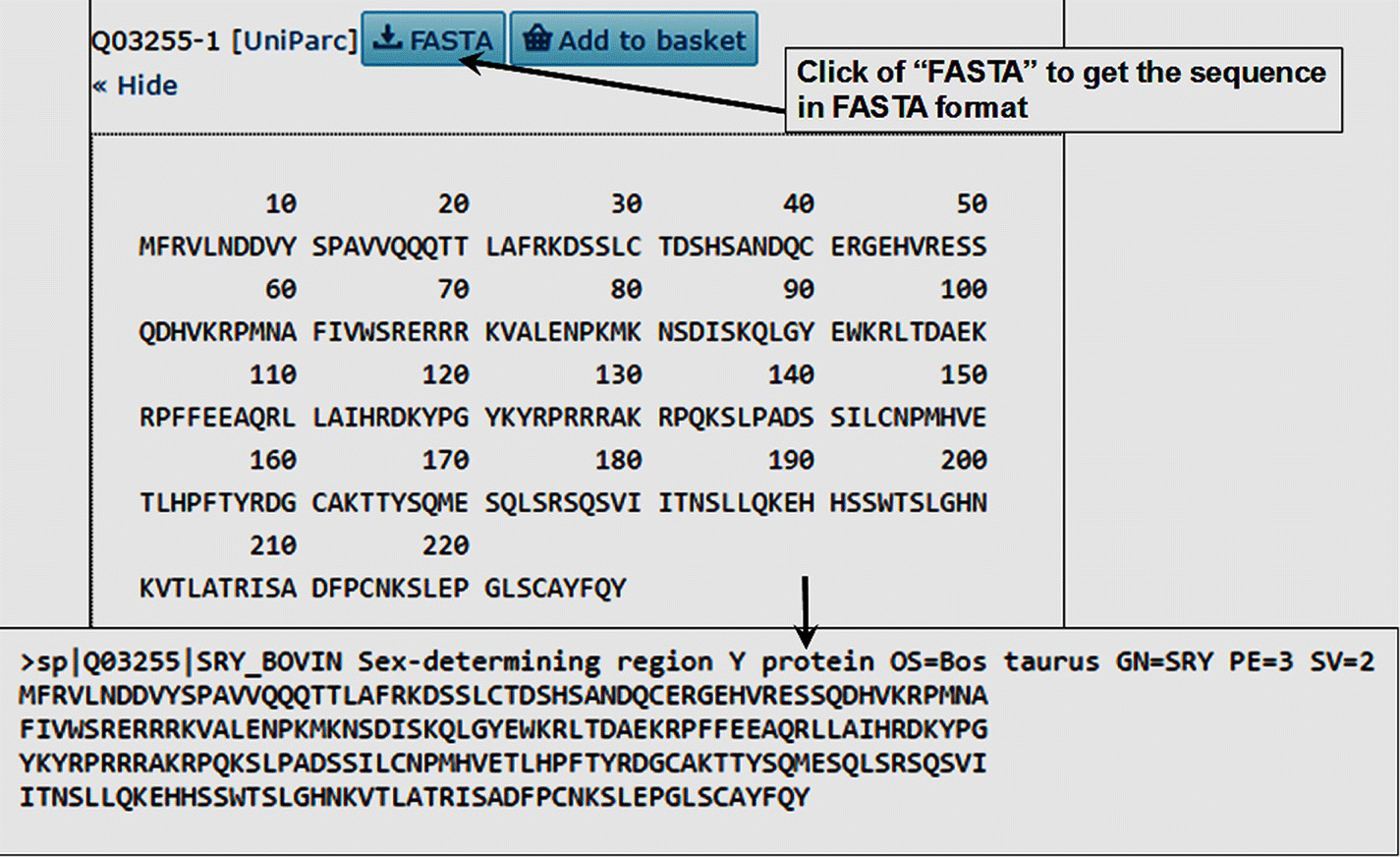
FIGURE 2.3 Peptide sequence of taurine SRY in FASTA format.
One can also BLAST the sequence or do the computation of physical as well as chemical parameters of the protein being studied (by ProtParam, i.e., Protein Parameters), compute pI/MW ratio and peptide mass to explore the molecular features of the protein.
Use the “Align” tab to align the above entry to its isoform (if the isoform is available). Note that the current entry (Q03255 (SRY_BOVIN)) does not have any isoform, so alignment is not possible.
The “Add to Basket Tab” is available to enable the user to select the entry and place it in a separate place (called “Basket”) for later use.
The “History” tab is meant for checking the history (dates of initial version, revised version, etc.) of entry.
2.4 QUESTIONS
- 1. Download the amino acid sequence of the Human TSPY protein from UniProtKB.
- 2. Write down the protein feature of the bovine SRY‐HMG‐box, using the ProtParam tool.
- 3. Enumerate the uses of UniProtKB/Swiss‐Prot vis‐à‐vis UniProtKB/TrEMBL.
- 4. Use the following sequence to find out the name of the protein (NCBI Protein Accession Number NP_001032554.1):
M A A A D G D D S L Y P I A V L I D E L R N E D V Q L R L N S I K K L S T I A L A L G V E R T R S E L L P F L T D T I Y D E D E V L L A L A E Q L G T F T T L V G G P E Y V H C L L P P L E S L A T V E E T V V R D K A V E S L R A I S H E H S P S D L E A H F V P L V K R L A G G D W F T S R T S A C G L F S V C Y P R V S S A V K A E L R Q Y F R N L C S D D T P M V R R A A A S K L G E F A K V L E L D N V K S E I I P M F S N L A S D E Q D S V R L L A V E A C V N I A Q L L P Q E D L E A L V M P T L R Q A A E D K S W R V R Y M V A D K F T E L H K A V G P E I T K T D L V P A F Q N L M K D C E A E V R A A A S H K V K E F C E N L S A D C R E N V I M T Q I L P C I K E L V S D A N Q H V K S A L A S V I M G L S P I L G K D S T I E H L L P L F L A Q L K D E C P E V R L N I I S N L D C V N E V I G I R Q L S Q S L L P A I V E L A E D A K W R V R L A I I E Y M P L L A G Q L G V E F F D E K L N S L C M A W L V D H V Y A I R E A A T S N L K K L V E K F G K E W A H A T I I P K V L A M S G D P N Y L H R M T T L F C I N V L S E V C G Q D I T T K H M L P T V L R M A G D P V A N V R F N V A K S L Q K I G P I L D N S T L Q S E V K P V L E K L T Q D Q D V D V K Y F A Q E A L T V L S L A
What is the SwissProt‐Id of the above sequence? Write down the common name of the protein, gene encoding the protein, and molecular weight of the protein.
- 5. Compare the above protein for the available information in NCBI (Protein Accession Number NP_001032554.1) and SwissProt (UniProtKB Id Q03255) databases.
