This section will provide a working example of the Apache Spark MLlib Naïve Bayes algorithm. It will describe the theory behind the algorithm, and will provide a step-by-step example in Scala to show how the algorithm may be used.
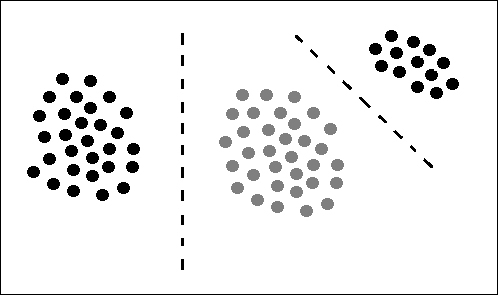
In order to use the Naïve Bayes algorithm to classify a data set, the data must be linearly divisible, that is, the classes within the data must be linearly divisible by class boundaries. The following figure visually explains this with three data sets, and two class boundaries shown via the dotted lines:
Naïve Bayes assumes that the features (or dimensions) within a data set are independent of one another, that is, they have no effect on each other. An example for Naïve Bayes is supplied with the help of Hernan Amiune at http://hernan.amiune.com/. The following example considers the classification of emails as spam. If you have 100 e-mails then perform the following:
60% of emails are spam 80% of spam emails contain the word buy 20% of spam emails don't contain the word buy 40% of emails are not spam 10% of non spam emails contain the word buy 90% of non spam emails don't contain the word buy
Thus, convert this example into probabilities, so that a Naïve Bayes equation can be created.
P(Spam) = the probability that an email is spam = 0.6 P(Not Spam) = the probability that an email is not spam = 0.4 P(Buy|Spam) = the probability that an email that is spam has the word buy = 0.8 P(Buy|Not Spam) = the probability that an email that is not spam has the word buy = 0.1
So, what is the probability that an e-mail that contains the word buy is spam? Well, this would be written as P (Spam|Buy). Naïve Bayes says that it is described by the equation in the following figure:

So, using the previous percentage figures, we get the following:
P(Spam|Buy) = ( 0.8 * 0.6 ) / (( 0.8 * 0.6 ) + ( 0.1 * 0.4 ) ) = ( .48 ) / ( .48 + .04 ) = .48 / .52 = .923
This means that it is 92 percent more likely that an e-mail that contains the word buy is spam. That was a look at the theory; now, it's time to try a real world example using the Apache Spark MLlib Naïve Bayes algorithm.
The first step is to choose some data that will be used for classification. I have chosen some data from the UK government data web site, available at: http://data.gov.uk/dataset/road-accidents-safety-data.
The data set is called "Road Safety - Digital Breath Test Data 2013," which downloads a zipped text file called DigitalBreathTestData2013.txt. This file contains around half a million rows. The data looks like this:
Reason,Month,Year,WeekType,TimeBand,BreathAlcohol,AgeBand,Gender Suspicion of Alcohol,Jan,2013,Weekday,12am-4am,75,30-39,Male Moving Traffic Violation,Jan,2013,Weekday,12am-4am,0,20-24,Male Road Traffic Collision,Jan,2013,Weekend,12pm-4pm,0,20-24,Female
In order to classify the data, I have modified both the column layout, and the number of columns. I have simply used Excel to give the data volume. However, if my data size had been in the big data range, I would have had to use Scala, or perhaps a tool like Apache Pig. As the following commands show, the data now resides on HDFS, in the directory named /data/spark/nbayes. The file name is called DigitalBreathTestData2013- MALE2.csv. Also, the line count from the Linux wc command shows that there are 467,000 rows. Finally, the following data sample shows that I have selected the columns: Gender, Reason, WeekType, TimeBand, BreathAlcohol, and AgeBand to classify. I will try and classify on the Gender column using the other columns as features:
[hadoop@hc2nn ~]$ hdfs dfs -cat /data/spark/nbayes/DigitalBreathTestData2013-MALE2.csv | wc -l 467054 [hadoop@hc2nn ~]$ hdfs dfs -cat /data/spark/nbayes/DigitalBreathTestData2013-MALE2.csv | head -5 Male,Suspicion of Alcohol,Weekday,12am-4am,75,30-39 Male,Moving Traffic Violation,Weekday,12am-4am,0,20-24 Male,Suspicion of Alcohol,Weekend,4am-8am,12,40-49 Male,Suspicion of Alcohol,Weekday,12am-4am,0,50-59 Female,Road Traffic Collision,Weekend,12pm-4pm,0,20-24
The Apache Spark MLlib classification functions use a data structure called LabeledPoint, which is a general purpose data representation defined at: http://spark.apache.org/docs/1.0.0/api/scala/index.html#org.apache.spark.mllib.regression.LabeledPoint.
This structure only accepts Double values, which means the text values in the previous data need to be classified numerically. Luckily, all of the columns in the data will convert to numeric categories, and I have provided two programs in the software package with this book, under the directory chapter2
aive bayes to do just that. The first is called convTestData.pl, and is a Perl script to convert the previous text file into Linux. The second file, which will be examined here is called convert.scala. It takes the contents of the DigitalBreathTestData2013- MALE2.csv file and converts each record into a Double vector.
The directory structure and files for an sbt Scala-based development environment have already been described earlier. I am developing my Scala code on the Linux server hc2nn using the Linux account hadoop. Next, the Linux pwd and ls commands show my top level nbayes development directory with the bayes.sbt configuration file, whose contents have already been examined:
[hadoop@hc2nn nbayes]$ pwd /home/hadoop/spark/nbayes [hadoop@hc2nn nbayes]$ ls bayes.sbt target project src
The Scala code to run the Naïve Bayes example is shown next, in the src/main/scala subdirectory, under the nbayes directory:
[hadoop@hc2nn scala]$ pwd /home/hadoop/spark/nbayes/src/main/scala [hadoop@hc2nn scala]$ ls bayes1.scala convert.scala
We will examine the bayes1.scala file later, but first, the text-based data on HDFS must be converted into the numeric Double values. This is where the convert.scala file is used. The code looks like this:
import org.apache.spark.SparkContext import org.apache.spark.SparkContext._ import org.apache.spark.SparkConf
These lines import classes for Spark context, the connection to the Apache Spark cluster, and the Spark configuration. The object that is being created is called convert1. It is an application, as it extends the class App:
object convert1 extends App {
The next line creates a function called enumerateCsvRecord. It has a parameter called colData, which is an array of strings, and returns a string:
def enumerateCsvRecord( colData:Array[String]): String = {
The function then enumerates the text values in each column, so for an instance, Male becomes 0. These numeric values are stored in values like colVal1:
val colVal1 = colData(0) match { case "Male" => 0 case "Female" => 1 case "Unknown" => 2 case _ => 99 } val colVal2 = colData(1) match { case "Moving Traffic Violation" => 0 case "Other" => 1 case "Road Traffic Collision" => 2 case "Suspicion of Alcohol" => 3 case _ => 99 } val colVal3 = colData(2) match { case "Weekday" => 0 case "Weekend" => 0 case _ => 99 } val colVal4 = colData(3) match { case "12am-4am" => 0 case "4am-8am" => 1 case "8am-12pm" => 2 case "12pm-4pm" => 3 case "4pm-8pm" => 4 case "8pm-12pm" => 5 case _ => 99 } val colVal5 = colData(4) val colVal6 = colData(5) match { case "16-19" => 0 case "20-24" => 1 case "25-29" => 2 case "30-39" => 3 case "40-49" => 4 case "50-59" => 5 case "60-69" => 6 case "70-98" => 7 case "Other" => 8 case _ => 99 }
A comma separated string called lineString is created from the numeric column values, and is then returned. The function closes with the final brace character}. Note that the data line created next starts with a label value at column one, and is followed by a vector, which represents the data. The vector is space separated while the label is separated from the vector by a comma. Using these two separator types allows me to process both: the label and the vector in two simple steps later:
val lineString = colVal1+","+colVal2+" "+colVal3+" "+colVal4+" "+colVal5+" "+colVal6 return lineString }
The main script defines the HDFS server name and path. It defines the input file, and the output path in terms of these values. It uses the Spark URL and application name to create a new configuration. It then creates a new context or connection to Spark using these details:
val hdfsServer = "hdfs://hc2nn.semtech-solutions.co.nz:8020" val hdfsPath = "/data/spark/nbayes/" val inDataFile = hdfsServer + hdfsPath + "DigitalBreathTestData2013-MALE2.csv" val outDataFile = hdfsServer + hdfsPath + "result" val sparkMaster = "spark://hc2nn.semtech-solutions.co.nz:7077" val appName = "Convert 1" val sparkConf = new SparkConf() sparkConf.setMaster(sparkMaster) sparkConf.setAppName(appName) val sparkCxt = new SparkContext(sparkConf)
The CSV-based raw data file is loaded from HDFS using the Spark context textFile method. Then, a data row count is printed:
val csvData = sparkCxt.textFile(inDataFile) println("Records in : "+ csvData.count() )
The CSV raw data is passed line by line to the enumerateCsvRecord function. The returned string-based numeric data is stored in the enumRddData variable:
val enumRddData = csvData.map { csvLine => val colData = csvLine.split(',') enumerateCsvRecord(colData) }
Finally, the number of records in the enumRddData variable is printed, and the enumerated data is saved to HDFS:
println("Records out : "+ enumRddData.count() ) enumRddData.saveAsTextFile(outDataFile) } // end object
In order to run this script as an application against Spark, it must be compiled. This is carried out with the sbt package command, which also compiles the code. The following command was run from the nbayes directory:
[hadoop@hc2nn nbayes]$ sbt package Loading /usr/share/sbt/bin/sbt-launch-lib.bash .... [info] Done packaging. [success] Total time: 37 s, completed Feb 19, 2015 1:23:55 PM
This causes the compiled classes that are created to be packaged into a JAR library, as shown here:
[hadoop@hc2nn nbayes]$ pwd /home/hadoop/spark/nbayes [hadoop@hc2nn nbayes]$ ls -l target/scala-2.10 total 24 drwxrwxr-x 2 hadoop hadoop 4096 Feb 19 13:23 classes -rw-rw-r-- 1 hadoop hadoop 17609 Feb 19 13:23 naive-bayes_2.10-1.0.jar
The application
convert1 can now be run against Spark using the application name, the Spark URL, and the full path to the JAR file that was created. Some extra parameters specify memory and maximum cores that are supposed to be used:
spark-submit --class convert1 --master spark://hc2nn.semtech-solutions.co.nz:7077 --executor-memory 700M --total-executor-cores 100 /home/hadoop/spark/nbayes/target/scala-2.10/naive-bayes_2.10-1.0.jar
This creates a data directory on HDFS called the /data/spark/nbayes/ followed by the result, which contains part files, containing the processed data:
[hadoop@hc2nn nbayes]$ hdfs dfs -ls /data/spark/nbayes Found 2 items -rw-r--r-- 3 hadoop supergroup 24645166 2015-01-29 21:27 /data/spark/nbayes/DigitalBreathTestData2013-MALE2.csv drwxr-xr-x - hadoop supergroup 0 2015-02-19 13:36 /data/spark/nbayes/result [hadoop@hc2nn nbayes]$ hdfs dfs -ls /data/spark/nbayes/result Found 3 items -rw-r--r-- 3 hadoop supergroup 0 2015-02-19 13:36 /data/spark/nbayes/result/_SUCCESS -rw-r--r-- 3 hadoop supergroup 2828727 2015-02-19 13:36 /data/spark/nbayes/result/part-00000 -rw-r--r-- 3 hadoop supergroup 2865499 2015-02-19 13:36 /data/spark/nbayes/result/part-00001
In the following HDFS cat command, I have concatenated the part file data into a file called DigitalBreathTestData2013-MALE2a.csv. I have then examined the top five lines of the file using the head command to show that it is numeric. Finally, I have loaded it into HDFS with the put command:
[hadoop@hc2nn nbayes]$ hdfs dfs -cat /data/spark/nbayes/result/part* > ./DigitalBreathTestData2013-MALE2a.csv [hadoop@hc2nn nbayes]$ head -5 DigitalBreathTestData2013-MALE2a.csv 0,3 0 0 75 3 0,0 0 0 0 1 0,3 0 1 12 4 0,3 0 0 0 5 1,2 0 3 0 1 [hadoop@hc2nn nbayes]$ hdfs dfs -put ./DigitalBreathTestData2013-MALE2a.csv /data/spark/nbayes
The following HDFS ls command now shows the numeric data file stored on HDFS, in the nbayes directory:
[hadoop@hc2nn nbayes]$ hdfs dfs -ls /data/spark/nbayes Found 3 items -rw-r--r-- 3 hadoop supergroup 24645166 2015-01-29 21:27 /data/spark/nbayes/DigitalBreathTestData2013-MALE2.csv -rw-r--r-- 3 hadoop supergroup 5694226 2015-02-19 13:39 /data/spark/nbayes/DigitalBreathTestData2013-MALE2a.csv drwxr-xr-x - hadoop supergroup 0 2015-02-19 13:36 /data/spark/nbayes/result
Now that the data has been converted into a numeric form, it can be processed with the MLlib Naïve Bayes algorithm; this is what the Scala file bayes1.scala does. This file imports the same configuration and context classes as before. It also imports MLlib classes for Naïve Bayes, vectors, and the LabeledPoint structure. The application class that is created this time is called bayes1:
import org.apache.spark.SparkContext import org.apache.spark.SparkContext._ import org.apache.spark.SparkConf import org.apache.spark.mllib.classification.NaiveBayes import org.apache.spark.mllib.linalg.Vectors import org.apache.spark.mllib.regression.LabeledPoint object bayes1 extends App {
Again, the HDFS data file is defined, and a Spark context is created as before:
val hdfsServer = "hdfs://hc2nn.semtech-solutions.co.nz:8020" val hdfsPath = "/data/spark/nbayes/" val dataFile = hdfsServer+hdfsPath+"DigitalBreathTestData2013-MALE2a.csv" val sparkMaster = "spark://hc2nn.semtech-solutions.co.nz:7077" val appName = "Naive Bayes 1" val conf = new SparkConf() conf.setMaster(sparkMaster) conf.setAppName(appName) val sparkCxt = new SparkContext(conf)
The raw CSV data is loaded and split by the separator characters. The first column becomes the label (Male/Female) that the data will be classified upon. The final columns separated by spaces become the classification features:
val csvData = sparkCxt.textFile(dataFile) val ArrayData = csvData.map { csvLine => val colData = csvLine.split(',') LabeledPoint(colData(0).toDouble, Vectors.dense(colData(1).split(' ').map(_.toDouble))) }
The data is then randomly divided into training (70%) and testing (30%) data sets:
val divData = ArrayData.randomSplit(Array(0.7, 0.3), seed = 13L) val trainDataSet = divData(0) val testDataSet = divData(1)
The Naïve Bayes MLlib function can now be trained using the previous training set. The trained Naïve Bayes model, held in the variable nbTrained, can then be used to predict the Male/Female result labels against the testing data:
val nbTrained = NaiveBayes.train(trainDataSet) val nbPredict = nbTrained.predict(testDataSet.map(_.features))
Given that all of the data already contained labels, the original and predicted labels for the test data can be compared. An accuracy figure can then be computed to determine how accurate the predictions were, by comparing the original labels with the prediction values:
val predictionAndLabel = nbPredict.zip(testDataSet.map(_.label)) val accuracy = 100.0 * predictionAndLabel.filter(x => x._1 == x._2).count() / testDataSet.count() println( "Accuracy : " + accuracy ); }
So this explains the Scala Naïve Bayes code example. It's now time to run the compiled bayes1 application using spark-submit, and to determine the classification accuracy. The parameters are the same. It's just the class name that has changed:
spark-submit --class bayes1 --master spark://hc2nn.semtech-solutions.co.nz:7077 --executor-memory 700M --total-executor-cores 100 /home/hadoop/spark/nbayes/target/scala-2.10/naive-bayes_2.10-1.0.jar
The resulting accuracy given by the Spark cluster is just 43 percent, which seems to imply that this data is not suitable for Naïve Bayes:
Accuracy: 43.30
In the next example, I will use K-Means to try and determine what clusters exist within the data. Remember, Naïve Bayes needs the data classes to be linearly divisible along the class boundaries. With K-Means, it will be possible to determine both: the membership and centroid location of the clusters within the data.
