This example will use the same test data from the previous example, but will attempt to find clusters in the data using the MLlib K-Means algorithm.
The K-Means algorithm iteratively attempts to determine clusters within the test data by minimizing the distance between the mean value of cluster center vectors, and the new candidate cluster member vectors. The following equation assumes data set members that range from X1 to Xn; it also assumes K cluster sets that range from S1 to Sk where K <= n.
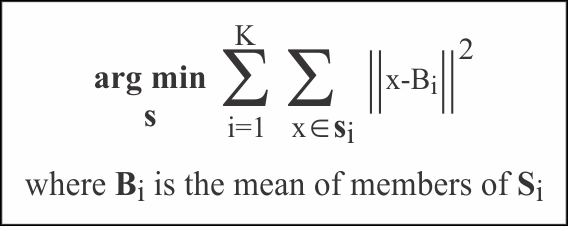
Again, the K-Means MLlib functionality uses the LabeledPoint structure to process its data and so, it needs numeric input data. As the same data from the last section is being reused, I will not re-explain the data conversion. The only change that has been made in data terms, in this section, is that processing under HDFS will now take place under the /data/spark/kmeans/ directory. Also, the conversion Scala script for the K-Means example produces a record that is all comma separated.
The development and processing for the K-Means example has taken place under the /home/hadoop/spark/kmeans directory, to separate the work from other development. The sbt configuration file is now called kmeans.sbt, and is identical to the last example, except for the project name:
name := "K-Means"
The code for this section can be found in the software package under chapter2K-Means. So, looking at the code for kmeans1.scala, which is stored under kmeans/src/main/scala, some similar actions occur. The import statements refer to Spark context and configuration. This time, however, the K-Means functionality is also being imported from MLlib. Also, the application class name has been changed for this example to kmeans1:
import org.apache.spark.SparkContext import org.apache.spark.SparkContext._ import org.apache.spark.SparkConf import org.apache.spark.mllib.linalg.Vectors import org.apache.spark.mllib.clustering.{KMeans,KMeansModel} object kmeans1 extends App {
The same actions are being taken as the last example to define the data file—define the Spark configuration and create a Spark context:
val hdfsServer = "hdfs://hc2nn.semtech-solutions.co.nz:8020" val hdfsPath = "/data/spark/kmeans/" val dataFile = hdfsServer + hdfsPath + "DigitalBreathTestData2013-MALE2a.csv" val sparkMaster = "spark://hc2nn.semtech-solutions.co.nz:7077" val appName = "K-Means 1" val conf = new SparkConf() conf.setMaster(sparkMaster) conf.setAppName(appName) val sparkCxt = new SparkContext(conf)
Next, the CSV data is loaded from the data file, and is split by comma characters into the variable VectorData:
val csvData = sparkCxt.textFile(dataFile) val VectorData = csvData.map { csvLine => Vectors.dense( csvLine.split(',').map(_.toDouble)) }
A K-Means object is initialized, and the parameters are set to define the number of clusters, and the maximum number of iterations to determine them:
val kMeans = new KMeans val numClusters = 3 val maxIterations = 50
Some default values are defined for initialization mode, the number of runs, and Epsilon, which I needed for the K-Means call, but did not vary for processing. Finally, these parameters were set against the K-Means object:
val initializationMode = KMeans.K_MEANS_PARALLEL val numRuns = 1 val numEpsilon = 1e-4 kMeans.setK( numClusters ) kMeans.setMaxIterations( maxIterations ) kMeans.setInitializationMode( initializationMode ) kMeans.setRuns( numRuns ) kMeans.setEpsilon( numEpsilon )
I cached the training vector data to improve the performance, and trained the K-Means object using the Vector Data to create a trained K-Means model:
VectorData.cache val kMeansModel = kMeans.run( VectorData )
I have computed the K-Means cost, the number of input data rows, and output the results via print line statements. The cost value indicates how tightly the clusters are packed, and how separated clusters are:
val kMeansCost = kMeansModel.computeCost( VectorData ) println( "Input data rows : " + VectorData.count() ) println( "K-Means Cost : " + kMeansCost )
Next, I have used the K-Means Model to print the cluster centers as vectors for each of the three clusters that were computed:
kMeansModel.clusterCenters.foreach{ println }
Finally, I have used the K-Means Model predict function to create a list of cluster membership predictions. I have then counted these predictions by value to give a count of the data points in each cluster. This shows which clusters are bigger, and if there really are three clusters:
val clusterRddInt = kMeansModel.predict( VectorData ) val clusterCount = clusterRddInt.countByValue clusterCount.toList.foreach{ println } } // end object kmeans1
So, in order to run this application, it must be compiled and packaged from the kmeans subdirectory as the Linux pwd command shows here:
[hadoop@hc2nn kmeans]$ pwd /home/hadoop/spark/kmeans [hadoop@hc2nn kmeans]$ sbt package Loading /usr/share/sbt/bin/sbt-launch-lib.bash [info] Set current project to K-Means (in build file:/home/hadoop/spark/kmeans/) [info] Compiling 2 Scala sources to /home/hadoop/spark/kmeans/target/scala-2.10/classes... [info] Packaging /home/hadoop/spark/kmeans/target/scala-2.10/k-means_2.10-1.0.jar ... [info] Done packaging. [success] Total time: 20 s, completed Feb 19, 2015 5:02:07 PM
Once this packaging is successful, I check HDFS to ensure that the test data is ready. As in the last example, I converted my data to numeric form using the convert.scala file, provided in the software package. I will process the data file DigitalBreathTestData2013-MALE2a.csv in the HDFS directory /data/spark/kmeans shown here:
[hadoop@hc2nn nbayes]$ hdfs dfs -ls /data/spark/kmeans Found 3 items -rw-r--r-- 3 hadoop supergroup 24645166 2015-02-05 21:11 /data/spark/kmeans/DigitalBreathTestData2013-MALE2.csv -rw-r--r-- 3 hadoop supergroup 5694226 2015-02-05 21:48 /data/spark/kmeans/DigitalBreathTestData2013-MALE2a.csv drwxr-xr-x - hadoop supergroup 0 2015-02-05 21:46 /data/spark/kmeans/result
The spark-submit tool is used to run the K-Means application. The only change in this command, as shown here, is that the class is now kmeans1:
spark-submit --class kmeans1 --master spark://hc2nn.semtech-solutions.co.nz:7077 --executor-memory 700M --total-executor-cores 100 /home/hadoop/spark/kmeans/target/scala-2.10/k-means_2.10-1.0.jar
The output from the Spark cluster run is shown to be as follows:
Input data rows : 467054 K-Means Cost : 5.40312223450789E7
The previous output shows the input data volume, which looks correct, plus it also shows the K-Means cost value. Next comes the three vectors, which describe the data cluster centers with the correct number of dimensions. Remember that these cluster centroid vectors will have the same number of columns as the original vector data:
[0.24698249738061878,1.3015883142472253,0.005830116872250263,2.9173747788555207,1.156645130895448,3.4400290524342454] [0.3321793984152627,1.784137241326256,0.007615970459266097,2.5831987075928917,119.58366028156011,3.8379106085083468] [0.25247226760684494,1.702510963969387,0.006384899819416975,2.231404248000688,52.202897927594805,3.551509158139135]
Finally, cluster membership is given for clusters 1 to 3 with cluster 1 (index 0) having the largest membership at 407,539 member vectors.
(0,407539) (1,12999) (2,46516)
So, these two examples show how data can be classified and clustered using Naïve Bayes and K-Means. But what if I want to classify images or more complex patterns, and use a black box approach to classification? The next section examines Spark-based classification using ANN's, or Artificial Neural Network's. In order to do this, I need to download the latest Spark code, and build a server for Spark 1.3, as it has not yet been formally released (at the time of writing).
