This chapter deals with evolutionary computing (EC) . I’ll start by defining EC and its scope so that you understand what is being discussed in this chapter and how it applies to AI. The following lists some of the major subtopics in the field of EC:
Evolutionary programming
Evolution strategies
Genetic algorithms
Genetic programming
Classifier systems
This list is by no means comprehensive, as the scope of many AI topics is often subject to the viewpoints of AI practitioners, but it serves me well and more than encompasses all the items discussed in this chapter.
I’ll begin with a story that should place the underlying EC concepts of evolution and mutation in a proper context.
Alife
Many years ago, there existed a large cavern in a subtropical jungle that was home to an amphibian creature, which I will simply call an alife. Alifes were gentle animals that made up a large colony (numbering in the tens of thousands) that lived in the cavern for a very long time. They ate lichen, moss, and other nutrients that were generously supplied by several clear water streams that flowed through the cavern. The alifes were quite prolific and bred very quickly, with generational times measured in weeks. Their colony size was also quite stable, basically set at an equilibrium point based on constant birth and death rates, and a stable food supply. All in all, the alife community seemed quite content with conditions as they were. Then a catastrophe struck.
A major earthquake shook the region, which was so strong that it collapsed the cavern roof, exposing the alifes to the outside world for the first time in eons. The alifes didn’t realize what had happened, having fairly primitive brains, and just continued with their ordinary ways. However, a hawk happened to be circling nearby and dove down through the newly opened hole to investigate. There it discovered the delicious alife colony and started consuming them. Having its fill, the hawk flew back to its nest area and communicated with other hawks about what it found. Very shortly, there were vast amounts of hungry hawks devouring the alifes. Things looked pretty grim for the alifes.
Fortunately, some groups of alifes were able to shelter in rocks and holes that the hawks could not reach. But it would only be a matter of time before the colony would be exterminated. Then, something remarkable happened. A skin cell on the top of the head of a newborn alife changed or mutated so that it became sensitive to light. The alife didn’t know what to do but primitive instincts directed it to avoid any light. This alife proceeded to have offspring and they too had the light-sensitive cells. Interestingly, each generation of alife became better at detecting light than the previous ones. These light-sensitive alifes quickly became somewhat adept at hiding from the hawks to the point that they were the only alifes that survived. The hawks left when it became apparent to them that the free lunch was over.
Another mutation happened with alifes, wherein a second set of light-sensitive cells developed fairly close to the first set. Over many generations, these light-sensitive cells evolved into primitive eyes. These alifes could now see, and more importantly, with two eyes they could perceive the third dimension of depth. With this new sense of depth, the alifes could venture forth and start exploring the world beyond their cavern home.
When the alifes started venturing from the cavern, they were exposed to the strong sunlight and their top skin cells started mutating, and became tougher and more protective. Their mouths also started mutating because they needed more food energy to move about in the jungle. They grew teeth and their digestive system changed to accommodate the raw meat proteins they were now consuming. Their bodies also started to grow to handle the new mass of organs and body parts. This evolution process continued for a very long time, until the present time. Today, there are alifes are among us, but we do not call them that; instead, we call them alligators.
I am sure that you recognize my story as fiction, except for the important parts of mutation and evolution that were necessary for the alifes to continue their species. The adaptation of species to survive in a hostile environment was the primary idea put forth in Charles Darwin’s thesis On the Origin of Species.
When it occurs in nature, mutation is always at a very small scale and usually based upon some random process. This idea is carried through in evolutionary computing, where changes or mutations are also very small and have little impact on the overall process, whatever that maybe. The mutations are also created by using a compatible pseudo-random mechanism. I’ll now further develop these ideas in a discussion of evolution programming (EP) .
Evolutionary Programing
EP was created by Dr. Lawrence Fogel in the early 1960s. It can be viewed as an optimization strategy using randomly selected trial solutions to test against one or more objectives. Trial solutions are also known as individual populations. Mutations are then applied to existing individuals, which create new individuals or offspring. The mutations can have a wide effect on the resultant behavior of the new individuals. New individuals are then compared in a “tournament” to select which ones survive to form a new individual population.
EP differs from a genetic algorithm (GA) because it focuses on the behavioral linkages between parents and offspring; whereas a GA tries to emulate nature’s genetic operations that occur in a genome, including encoding behaviors and recombinations using genetic crossovers.
EP is also very similar to evolution strategy (ES) , even though they were developed independently of each other. The main difference is that EP uses a random process to select individuals from a population, compared to a deterministic approach used in ES. In ES, poorly performing individuals are deleted from the individual population based on well-defined metrics.
Now that I have introduced EC and discussed its fundamental components, it is time to show you a practical EC demonstration.
Demo 10-1: Manual Calculation
I begin this demonstration with some manual calculations, as I have done in other chapters. However, it would be helpful to state the purpose so that you have an idea about what the demonstration is supposed to show. The purpose is to generate a list of six integers whose values range from 0 to 100 and whose sum is 371. I can guess that most readers can easily come up with a list without any real issues.
I will take you through my reasoning to illustrate how I developed a list.
First, I recognized that that each number is likely more than 60 because there are only six numbers available to sum to the target value.
Next, I selected a number (say 71) and subtracted it from the target, which caused a new target of 300 to be created with the five numbers left.
I repeated these steps using other numbers until I arrived at the following list. The last number was simply the remainder after I selected the fifth integer.
[71, 90, 65, 70, 25, 50]
This process was not randomized in any way as I reasoned my selections throughout the process. This process should be classified as deterministic. A traditional script or program could be written to codify it. Incidentally, I could have also created the following list because there was no stipulation that integers could not be repeated:
[60, 60, 60, 60, 60, 71]
It is just a quirk of human nature that we usually do not think or reason in that manner.
I would consider the preceding manual calculations as fairly trivial for a human. However, it is not so trivial for a computer, and this is where the following Python demonstration comes into play.
Python Script
Credit must go to Will Larson. I am using his code from a 2009 article entitled “Genetic Algorithms: Cool Name & Damn Simple” ( https://lethain.com/genetic-algorithms-cool-name-damn-simple/ ) from his blog, which is called Irrational Exuberance. I highly recommend that you take a look at it.
The problem to be solved is the same one used in the manual calculations section on determining six integers with values ranging from 0 to 100 and summing to a target value of 371.
The first item to consider in formulating a solution is how to structure it to fit into the EC paradigm. There are individuals to create who will eventually form a population. The individuals for this specific case will be six element lists consisting of integers whose values range from 0 to 100. Multiple individuals make up a population. The following code segment creates the individuals:
from random import randintdef individual(length, min, max):# generates an individualreturn [randint(min, max) for x in xrange(length)]
Figure 10-1 shows an interactive Python session where I generated several individuals.
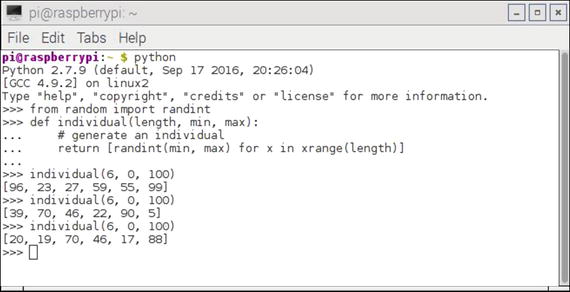
Figure 10-1. Interactive Python session individual generation
The individuals generated must be collected so that they form a population, which is the next part of the solution structure. The following code segment generates a population. This segment depends upon the previous code segment to have already been entered:
def population(count, length, min, max):# generate a populationreturn [individual(length, min, max) for x in xrange(count)]
Figure 10-2 shows an interactive Python session where I generated several populations.
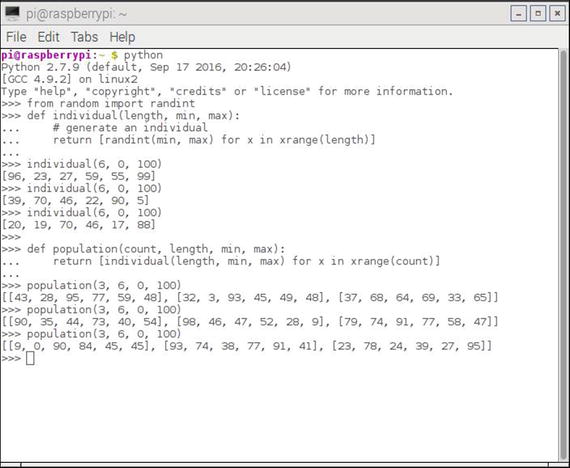
Figure 10-2. Interactive Python session population generation
The next step in this process is to create a function that measures how well a particular individual performs in meeting the stated objective (i.e., integer lists values summing to a target value). This function is called a fitness function. Note that the fitness function requires the individual function to have been previously entered. The following is a code segment implementation for the fitness function:
from operator import adddef fitness(individual, target):# calculate fitness, lower the bettersum = reduce(add, individual, 0)return abs(target - sum)
Figure 10-3 shows an interactive Python session where I tested several individuals against a constant target value.
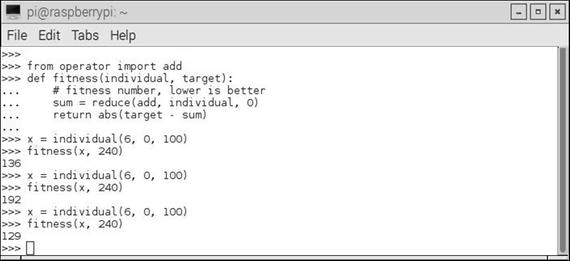
Figure 10-3. Interactive Python session fitness tests
This particular fitness function is much simpler than similar fitness tests that I have demonstrated in past chapters. In this one, only the absolute value of the difference between the summed elements contained in an individual list and a target value is calculated. The best case would be a 0.0 value, which I will shortly demonstrate.
The only missing element in the structure is how to change or evolve the population to meet the objective. Only by the purest luck would an initial solution also be an optimal solution. There must be a strategy stated to properly implement an evolutionary function. The following is the strategy set forth for this structure :
Take 20% of the top performers (elitism rate) from a prior population and include them in a new one.
Breed approximately 75% of the new population as children.
Take the first length/2 elements from a father and the last length/2 elements from a mother to form a child.
It is forbidden to have a father and a mother as the same individual.
Randomly select 5% from the population.
Mutate 1% of the new population.
This strategy is by no means a standard one, or even a very comprehensive one, but it will suffice for this problem and it is fairly representative of those used for similar problems.
Figure 10-4 is an interactive Python session that shows how children are formed in this strategy.
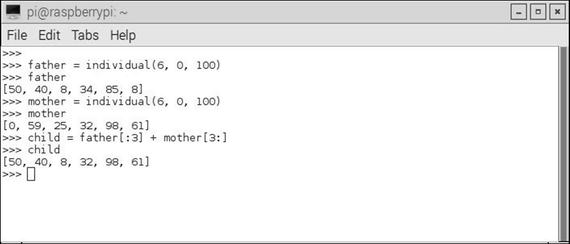
Figure 10-4. Forming children for a new population
The mutating part of the strategy is a bit more complex, which I show, in part, with the next code segment before trying to explain it:
from random import random, randintchance_to_mutate = 0.01for i in population:if chance_to_mutate > random():place_to_mutate = randint(0, len(i))i[place_to_mutate] = randint(min(i), max(i))......
The chance_to_mutate variable is set to 0.01, representing a 1% chance for mutation, which is very low as I noted earlier. The statement for i in population: iterates through the entire population and causes a mutation only when the random number generator is less than .01, which is not very often. The actual individual chosen to be mutated is accomplished by the place_to_mutate = randint(0, len(i)) statement, which is not likely the individual that happened to be iterated upon at the time the random number was less than .01. Finally, the actual mutation is done by this statement: i[place_to_mutate] = randint(min(i), max(i)). The integer values in the selected individual are randomly generated based upon the population’s min and max values.
The whole strategy design, which includes a mix of selecting the best-performing individuals, breeding children from all portions of the population, and the occasional mutations is geared toward finding the global maximum and avoiding local maximums. This is precisely the same thought process that was going on in the gradient descent algorithm for ANNs, where the goal was to locate a global minimum and avoid local minimums. You can look at Figure 8-15 to visualize the process for locating the global maximum or highest peak instead of the deepest valley, representing the global minimum.
There is much more to the evolve function than what was shown. The remaining code is shown next in the complete script listing.
There is one more function to explain before the complete script is shown. This function is named grade and it calculates an overall fitness measure for a whole population. The Python built-in reduce function sums the fitness scores for each individual and averages the sum by the population size. The following code implements the grade function :
def grade(pop, target):'Find average fitness for a population.'summed = reduce(add, (fitness(x, target) for x in pop))return summed / (len(pop) * 1.0)
This last function listing concludes my discussion on all the functions that compose the Python script. The following is a complete listing of the final script, along with instructions on how to run the script within an interactive Python session. Note that I did modify the instructions to display the first generation number that met the target and the final solution itself. The population in the example is equal to 100 and each individual has six elements with values ranging between 0 and 100.
"""# Example usage>>> from genetic import *>>> target = 371>>> p_count = 100>>> i_length = 6>>> i_min = 0>>> i_max = 100>>> p = population(p_count, i_length, i_min, i_max)>>> fitness_history = [grade(p, target),]>>> fitFlag = 0>>> for i in xrange(100):... p = evolve(p, target)... fitness_history.append(grade(p, target))... if grade(p, target) == 0:... if fitFlag == 0:... fitFlag = 1... print 'Generation = ', i... print p[0]>>> for datum in fitness_history:... print datum"""from random import randint, randomfrom operator import adddef individual(length, min, max):'Create a member of the population.'return [ randint(min,max) for x in xrange(length) ]def population(count, length, min, max):"""Create a number of individuals (i.e. a population).count: the number of individuals in the populationlength: the number of values per individualmin: the minimum possible value in an individual's list of valuesmax: the maximum possible value in an individual's list of values"""return [ individual(length, min, max) for x in xrange(count) ]def fitness(individual, target):"""Determine the fitness of an individual. Higher is better.individual: the individual to evaluatetarget: the target number individuals are aiming for"""sum = reduce(add, individual, 0)return abs(target-sum)def grade(pop, target):'Find average fitness for a population.'summed = reduce(add, (fitness(x, target) for x in pop))return summed / (len(pop) * 1.0)def evolve(pop, target, retain=0.2, random_select=0.05, mutate=0.01):graded = [ (fitness(x, target), x) for x in pop]graded = [ x[1] for x in sorted(graded)]retain_length = int(len(graded)*retain)parents = graded[:retain_length]# randomly add other individuals to# promote genetic diversityfor individual in graded[retain_length:]:if random_select > random():parents.append(individual)# mutate some individualsfor individual in parents:if mutate > random():pos_to_mutate = randint(0, len(individual)-1)# this mutation is not ideal, because it# restricts the range of possible values,# but the function is unaware of the min/max# values used to create the individuals,individual[pos_to_mutate] = randint(min(individual), max(individual))# crossover parents to create childrenparents_length = len(parents)desired_length = len(pop) - parents_lengthchildren = []while len(children) < desired_length:male = randint(0, parents_length-1)female = randint(0, parents_length-1)if male != female:male = parents[male]female = parents[female]half = len(male) / 2child = male[:half] + female[half:]children.append(child)parents.extend(children)return parents
Figure 10-5 shows an interactive session in which I entered all the statements shown in the instructions portion of the script comments.
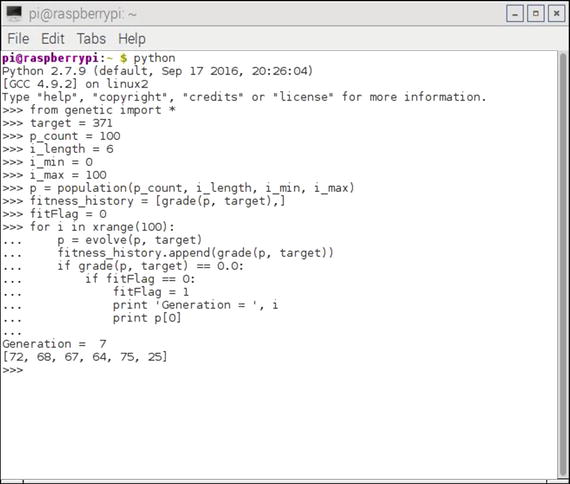
Figure 10-5. Interactive Python session running the script
You should be able to see in the screenshot that a solution was found after only seven generations had evolved. The solution was [72, 68, 67, 64, 75, 25], which does sum to the target value of 371. The script does not stop after the first successful solution is found, but continues to evolve and mutate, slightly degrading and then improving until it has run through its preset cycle number.
A portion of the history of the fitness numbers associated with each generation is shown in Figure 10-6.
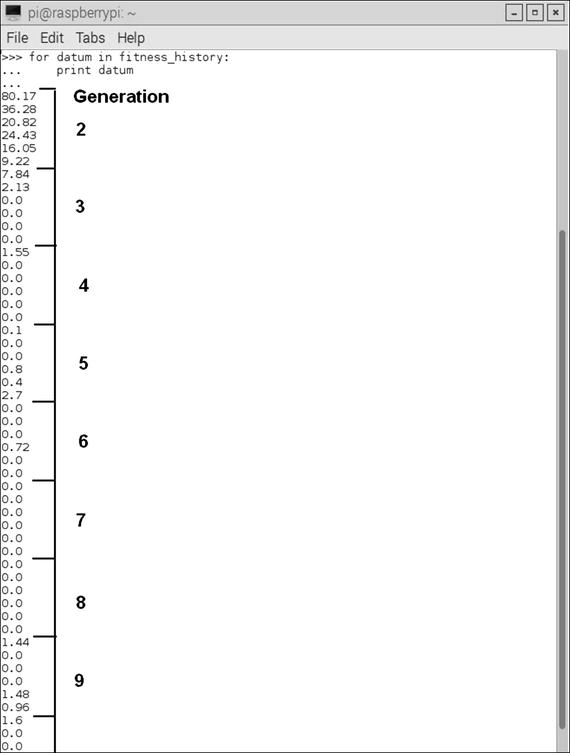
Figure 10-6. Fitness history list
In Figure 10-6, I annotated where generation 7 has a 0.0 fitness value for each of its individuals. Overall, this genetic programming approach is very efficient, especially for the relatively simple objective of determining a target sum given a list of six randomly generated integer numbers.
The next demonstration is a slight variant from Conway’s Game of Life, which is a classic project incorporating genetic programming with artificial life (alife) that breed and die according to a set of conditions based on their proximity to each other, which I explain later.
Demo 10-2: Conway’s Game of Life
The Game of Life, or as it is commonly known, Life, is a cellular automaton project created by British mathematician John Conway in 1970. The game starts with an initial condition, but it needs no further user input to play to its completion. This modus operandi is called a zero-player game, which means that the automatons—or cells, as I shall refer to them from now on—evolve on their own according to the following set of rules or conditions :
Any live cell with fewer than two live neighbors dies, as if caused by underpopulation.
Any live cell with two or three live neighbors lives to the next generation.
Any live cell with more than three live neighbors dies, as if by overpopulation.
Any dead cell with exactly three live neighbors becomes a live cell, as if by reproduction.
The board field, or universe, for this game is theoretically an infinite, orthogonal set of grid squares where a cell can occupy a grid in either an alive or a dead state. Alive is synonymous with populated and dead is synonymous with unpopulated. Each cell has a maximum of eight neighbors, except for the edge cells in our real-world, practical grid system, where an infinite grid is not possible.
The grid is “seeded” with an initial placement of cells, which can be randomly or deterministically placed. The cellular rules are then immediately applied, causing births and deaths to happen simultaneously. This application is known as time tick in game terminology. A new generation is thus formed and the rules are immediately reapplied. Ultimately, the game settles into an equilibrium state, where cells cycle between two stable cellular configurations, they roam about forever, or they all die.
From a historical perspective, Conway was very much interested in Professor John von Neumann’s attempts at creating a computing machine that could replicate itself. von Neumann eventually succeeded by describing a mathematical model based on a rectangular grid governed by a very complex set of rules. The Conway game was a big simplification of von Neumann’s concepts. The game was published in the October 1970 issue of Scientific American in Martin Gardener’s “Mathematical Games” column. It instantly became a huge success and generated much interest from fellow AI researchers and enthusiastic readers.
The game can also be extended to the point that it is comparable to the universal Turing machine, first proposed by Alan Turning in the 1940s (explained in an earlier chapter).
The game has another important influence on AI in that it likely kick-started the mathematical field of study known as cellular automata. The game simulates the birth and death of a colony of organisms in a surprising close relationship to real-life processes occurring in nature. This game has led directly to many other similar simulation games modeling nature’s own processes. These simulations have been applied in computer science, biology, physics, biochemistry, economics, mathematics, and many others.
I demonstrate Conway’s Game of Life using a neat Raspberry Pi accessory board known as a Sense HAT. Figure 10-7 shows a Sense HAT board. The HAT name represents a line of accessory boards known as Hardware Attached on Top (HAT) boards, which feature a standardized format that allows them to be directly plugged into the 40-pin GPIO header and mechanically fastened to the Raspberry Pi 2 and 3 model boards.
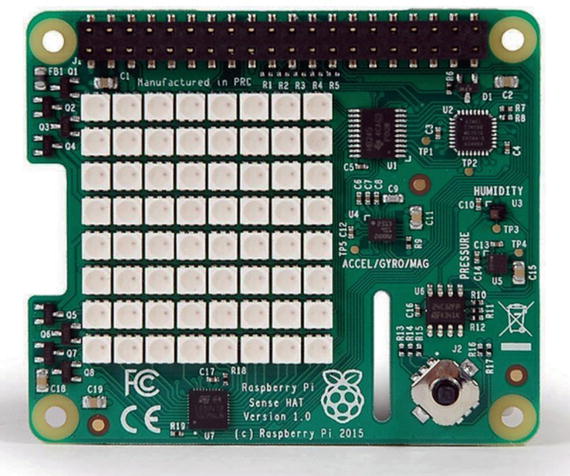
Figure 10-7. Sense HAT board
Every HAT board supports an autoconfiguration system that allows automatic GPIO and driver setup. This automatic configuration is achieved using two dedicated pins on the 40-pin GPIO header that are reserved for an I2C eeprom. This eeprom holds the board manufacturer’s information, GPIO setup, and a device tree fragment, which is a description of the attached hardware, which in turn allows the Linux OS to automatically load any required drivers.
The Sense HAT board has an 8 × 8 RGB LED array, which provides a very nice grid display for all the cellular automatons. In addition, there is a powerful Python library that provides a good deal of functionality for the LED array and for a variety of onboard sensors , which include the following:
Gyroscope
Accelerometer
Magnetometer
Temperature
Barometric pressure
Humidity
There is also a five-way position joystick on the board to support applications that need that type of user control. I do not use any of the sensors or the joystick for the Game of Life application, just the 8 × 8 LED array.
Sense HAT Hardware Installation
First, ensure that the Raspberry Pi is completely powered down. The Sense HAT is designed to mount on top of a Raspberry Pi. It comes with a 40-pin GPIO male pin extension header that you must first plug into the female 40-pin header on the Raspberry Pi. The Sense HAT is then mounted on top of the Raspberry Pi using the 40 male pins as a guide. These pins should simply push through the 40-pin female header on the Sense board. All that’s left is to attach the supplied stand-offs, which provide firm support between the Raspberry Pi and the Sense HAT. Figure 10-8 shows a mounted Sense HAT on a Raspberry Pi.
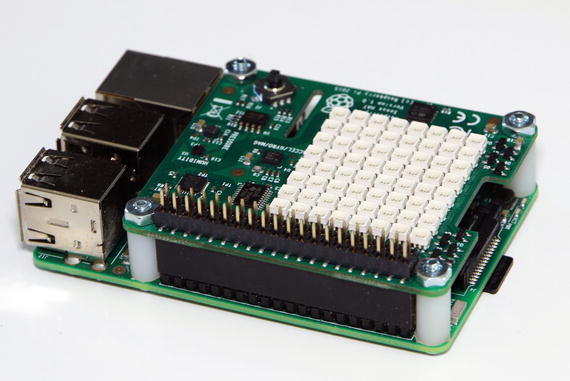
Figure 10-8. Sense HAT mounted onto a Raspberry Pi
There is one more item that you should know. The Raspberry Pi and Sense HAT require a power supply capable of providing 2.5A at 5V. Failing to use a sufficiently powerful power supply will likely cause strange behavior, such as the Linux OS failing to recognize the Sense HAT, which I unfortunately confirmed as a fact.
Sense HAT Software Installation
The Python library supporting the Sense HAT must be loaded prior to running any of the scripts that will run the Life game. First, ensure that the Raspberry Pi is connected to the Internet, and then enter the following commands to load the software:
sudo apt-get updatesudo apt-get install sense-hatsudo reboot
You want to run the following test to ensure that the Sense HAT is working properly with the newly installed software. Enter the following in an interactive Python session:
from sense_hat import SenseHatsense = SenseHat()sense.show_message("Hello World!")
If all went well with the installation, you show see the Hello World! message scrolling across the LED array. If you don’t see it, I would recheck that the Sense HAT is securely fastened to the Raspberry Pi and that all 40 pins go through their respective socket holes. It very well might be that you inadvertently bent one of the pins such that it is has been forced out of the way. If that’s the case, just carefully straighten it and reseat all the pins.
Once the software checks out, you are ready to run a Python version of the Game of Life on this combination of Sense HAT/Raspberry Pi. I now turn the discussion to the game software.
Game of Life: Python Version
I begin this section by giving credit to Mr. Swee Meng Ng, a very talented Malaysian developer who posted much of the code I used in this project on GitHub.com. Swee goes by the username sweemeng on GitHub. He has a blog at www.nomadiccodemonkey.com . Take a look at it if you want an appreciation for the code development efforts that are ongoing in that part of the world.
You should first load the following Python scripts from https://github.com/sweemeng/sweemengs-game-of-life.git into your home directory:
genelab.ppy
designer.py
gameoflife.py
genelab.py is the first script that I’ll discuss. It is the main one in this application. By main, I mean it incorporates the starting point, initialization, generation creation, and mutation, and finally, it enforces the behavioral rules. However, this script needs two helper scripts to work properly. These helper scripts are named designer.py and gameoflife.py, which I discuss shortly. I have added my own comments to Swee Meng’s scripts to help relate portions of it to concepts that have already been discussed, and hopefully, to clarify the purpose of the code segments.
import randomimport time# first helperfrom designer import CellDesignerfrom designer import GeneBank# second helperfrom gameoflife import GameOfLife# not exactly a helper but needed for the displayfrom sense_hat import SenseHat# you can customize your colors hereWHITE = [ 0, 0, 0 ]RED = [ 120, 0, 0 ]# begin class definitionclass Genelab(object):# begin initializationdef __init__(self):self.survive_min = 5 # Cycleself.surival_record = 0self.designer = CellDesigner()self.gene_bank = GeneBank()self.game = GameOfLife()self.sense = SenseHat()# random starting pointdef get_start_point(self):x = random.randint(0, 7)y = random.randint(0, 7)return x, y# create a fresh generation (population)# or mutate an existing onedef get_new_gen(self):if len(self.gene_bank.bank) == 0:print("creating new generation")self.designer.generate_genome()elif len(self.gene_bank.bank) == 1:print("Mutating first gen")self.designer.destroy()seq_x = self.gene_bank.bank[0]self.designer.mutate(seq_x)else:self.designer.destroy()coin_toss = random.choice([0, 1])if coin_toss:print("Breeding")seq_x = self.gene_bank.random_choice()seq_y = self.gene_bank.random_choice()self.designer.cross_breed(seq_x, seq_y)else:print("Mutating")seq_x = self.gene_bank.random_choice()self.designer.mutate(seq_x)# a method to start the whole game, i.e. lab.run()def run(self):self.get_new_gen()x, y = self.get_start_point()cells = self.designer.generate_cells(x, y)self.game.set_cells(cells)# count is the generation numbercount = 1self.game.destroy_world()# forever loop. Change this if you only want a finite runwhile True:try:# essentially where the rules are appliedif not self.game.everyone_alive():if count > self.survive_min:# Surivival the fittestself.gene_bank.add_gene(self.designer.genome)self.survival_record = countprint("Everyone died, making new gen")print("Species survived %s cycle" % count)self.sense.clear()self.get_new_gen()x, y = self.get_start_point()cells = self.designer.generate_cells(x, y)self.game.set_cells(cells)count = 1if count % random.randint(10, 100) == 0:print("let's spice thing up a little")print("destroying world")print("Species survived %s cycle" % count)self.game.destroy_world()self.gene_bank.add_gene(self.designer.genome)self.sense.clear()self.get_new_gen()x, y = self.get_start_point()cells = self.designer.generate_cells(x, y)self.game.set_cells(cells)count = 1canvas = []# this where the cells are "painted" onto the canvas# The canvas is based on the grid pattern from the# gameoflife scriptfor i in self.game.world:if not i:canvas.append(WHITE)else:canvas.append(RED)self.sense.set_pixels(canvas)self.game.run()count = count + 1time.sleep(0.1)except:print("Destroy world")print("%s generation tested" % len(self.gene_bank.bank))self.sense.clear()breakif __name__ == "__main__":# instantiate the class GeneLablab = Genelab()# start the gamelab.run()
The first helper script is designer.py. The code listing follows with the addition of my own comments:
import randomclass CellDesigner(object):# initializationdef __init__(self, max_point=7, max_gene_length=10, genome=[]):self.genome = genomeself.max_point = max_pointself.max_gene_length = max_gene_length# a genome is made up of 1 to 10 genesdef generate_genome(self):length = random.randint(1, self.max_gene_length)print(length)for l in range(length):gene = self.generate_gene()self.genome.append(gene)# a gene is an (+/-x, +/-y) cooordinate pair; x, y range 0 to 7def generate_gene(self):x = random.randint(0, self.max_point)y = random.randint(0, self.max_point)x_dir = random.choice([1, -1])y_dir = random.choice([1, -1])return ((x * x_dir), (y * y_dir))def generate_cells(self, x, y):cells = []for item in self.genome:x_move, y_move = itemnew_x = x + x_moveif new_x > self.max_point:new_x = new_x - self.max_pointif new_x < 0:new_x = self.max_point + new_xnew_y = y + x_moveif new_y > self.max_point:new_y = new_y - self.max_pointif new_y < 0:new_y = self.max_point + new_ycells.append((new_x, new_y))return cellsdef cross_breed(self, seq_x, seq_y):if len(seq_x) > len(seq_y):main_seq = seq_xsecondary_seq = seq_yelse:main_seq = seq_ysecondary_seq = seq_xfor i in range(len(main_seq)):gene = random.choice([ main_seq, secondary_seq ])if i > len(gene):continueself.genome.append(gene[i])def mutate(self, sequence):# Just mutate one genefor i in sequence:mutate = random.choice([ True, False ])if mutate:gene = self.generate_gene()self.genome.append(gene)else:self.genome.append(i)def destroy(self):self.genome = []class GeneBank(object):def __init__(self):self.bank = []def add_gene(self, sequence):self.bank.append(sequence)def random_choice(self):if not self.bank:return Nonereturn random.choice(self.bank)
The second helper script is gameoflife.py. There is only a small portion of this script that is actually used as a helper for the main script. I have included it all for the sake of completeness and to provide you with the code, in case you want to run a single-generation Game of Life, which I will explain shortly. The complete code listing follows with my own comments added:
import timeworld = [0, 0, 0, 0, 0, 0, 0, 0,0, 0, 0, 0, 0, 0, 0, 0,0, 0, 0, 0, 0, 0, 0, 0,0, 0, 0, 0, 0, 0, 0, 0,0, 0, 0, 0, 0, 0, 0, 0,0, 0, 0, 0, 0, 0, 0, 0,0, 0, 0, 0, 0, 0, 0, 0,0, 0, 0, 0, 0, 0, 0, 0,]max_point = 7 # We use a square world to make things easyclass GameOfLife(object):def __init__(self, world=world, max_point=max_point, value=1):self.world = worldself.max_point = max_pointself.value = valuedef to_reproduce(self, x, y):if not self.is_alive(x, y):neighbor_alive = self.neighbor_alive_count(x, y)if neighbor_alive == 3:return Truereturn Falsedef to_kill(self, x, y):if self.is_alive(x, y):neighbor_alive = self.neighbor_alive_count(x, y)if neighbor_alive < 2 or neighbor_alive > 3:return Truereturn Falsedef to_keep(self, x, y):if self.is_alive(x, y):neighbor_alive = self.neighbor_alive_count(x, y)if neighbor_alive >= 2 and neighbor_alive <= 3:return Truereturn Falsedef is_alive(self, x, y):pos = self.get_pos(x, y)return self.world[pos]def neighbor_alive_count(self, x, y):neighbors = self.get_neighbor(x, y)alives = 0for i, j in neighbors:if self.is_alive(i, j):alives = alives + 1# Because neighbor comes with self, just for easinessif self.is_alive(x, y):return alives - 1return alivesdef get_neighbor(self, x, y):#neighbors = [# (x + 1, y + 1), (x, y + 1), (x - 1, y + 1),# (x + 1, y), (x, y), (x, y + 1),# (x + 1, y - 1), (x, y - 1), (x - 1, y - 1),#]neighbors = [(x - 1, y - 1), (x - 1, y), (x - 1, y + 1),(x, y - 1), (x, y), (x, y + 1),(x + 1, y - 1), (x + 1, y), (x + 1, y + 1)]return neighborsdef get_pos(self, x, y):if x < 0:x = max_pointif x > max_point:x = 0if y < 0:y = max_pointif y > max_point:y = 0return (x * (max_point+1)) + y# I am seriously thinking of having multiple speciesdef set_pos(self, x, y):pos = self.get_pos(x, y)self.world[pos] = self.valuedef set_cells(self, cells):for x, y in cells:self.set_pos(x, y)def unset_pos(self, x, y):pos = self.get_pos(x, y)self.world[pos] = 0def run(self):something_happen = Falseoperations = []for i in range(max_point + 1):for j in range(max_point + 1):if self.to_keep(i, j):something_happen = Truecontinueif self.to_kill(i, j):operations.append((self.unset_pos, i, j))something_happen = Truecontinueif self.to_reproduce(i, j):something_happen = Trueoperations.append((self.set_pos, i, j))continuefor func, i, j in operations:func(i, j)if not something_happen:print("weird nothing happen")def print_world(self):count = 1for i in self.world:if count % 8 == 0:print("%s " %i)else:print("%s " %i) #, end = "")count = count + 1print(count)def print_neighbor(self, x, y):neighbors = self.get_neighbor(x, y)count = 1for i, j in neighbors:pos = self.get_pos(i, j)if count %3 == 0:print("%s " %self.world[pos])else:print("%s " %self.world[pos]) #, end = "")count = count + 1print(count)def everyone_alive(self):count = 0for i in self.world:if i:count = count + 1if count:return Truereturn Falsedef destroy_world(self):for i in range(len(self.world)):self.world[i] = 0def main():game = GameOfLife()cells = [ (2, 4), (3, 5), (4, 3), (4, 4), (4, 5) ]game.set_cells(cells)print(cells)while True:try:game.print_world()game.run()count = 0time.sleep(5)except KeyboardInterrupt:print("Destroy world")breakdef debug():game = GameOfLife()cells = [ (2, 4), (3, 5), (4, 3), (4, 4), (4, 5) ]game.set_cells(cells)test_cell = (3, 3)game.print_neighbor(*test_cell)print("Cell is alive: %s" % game.is_alive(*test_cell))print("Neighbor alive: %s" % game.neighbor_alive_count(*test_cell))print("Keep cell: %s" % game.to_keep(*test_cell))print("Make cell: %s" % game.to_reproduce(*test_cell))print("Kill cell: %s" % game.to_kill(*test_cell))game.print_world()game.run()game.print_world()if __name__ == "__main__":main()#debug()
Test Run
First, ensure that the genelab.py , designer.py, and gameoflife.py scripts are in the pi home directory before running this command:
python genelab.pyThe Raspberry takes a moment to load everything. You should begin to see the cells appear on the Sense HAT LED array, as well as status messages on the console screen. Figure 10-9 is a photograph of the LED array while my script was running.
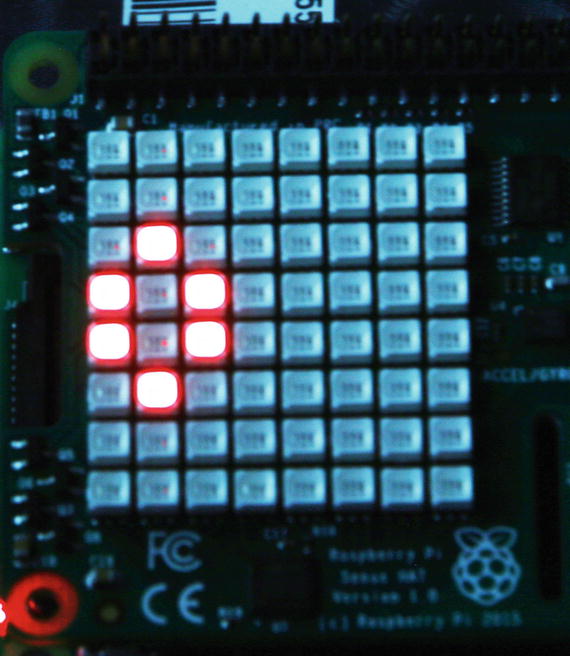
Figure 10-9. Sense HAT LED array with the Game of Life running
Figure 10-10 shows the console display while the game was running.
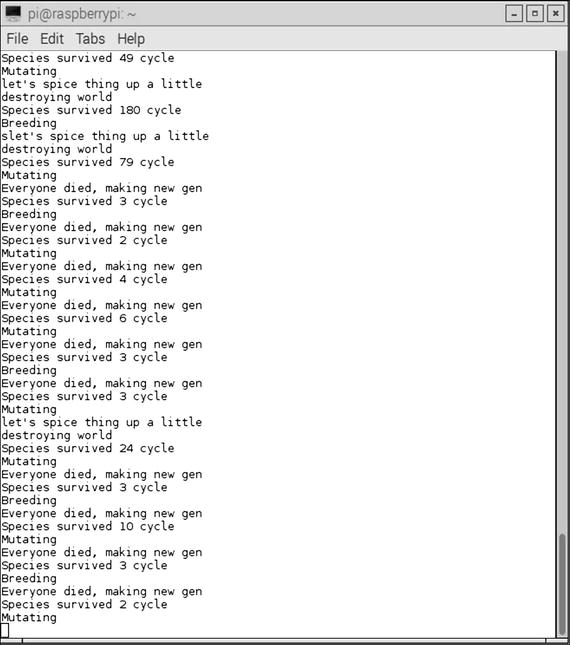
Figure 10-10. Console display with the Game of Life running
Single Generation of the Game of Life
It is also entirely possible to experiment with only a single generation of the Game of Life. This script simply adheres to the rules for the cell neighbors that were previously specified, with no cell or gene mutations allowed. The following script is named main.py. It is available on the same GitHub website as the previous scripts.
from sense_hat import SenseHatfrom gameoflife import GameOfLifeimport timeWHITE = [ 0, 0, 0 ]RED = [ 255, 0, 0 ]def main():game = GameOfLife()sense = SenseHat()# cells = [(2, 4), (3, 5), (4, 3), (4, 4), (4, 5)]cells = [(2, 4), (2, 5), (1,5 ), (1, 6), (3, 5)]game.set_cells(cells)while True:try:canvas = []for i in game.world:if not i:canvas.append(WHITE)else:canvas.append(RED)sense.set_pixels(canvas)game.run()if not game.everyone_alive():sense.clear()print("everyone died")breaktime.sleep(0.1)except:sense.clear()breakif __name__ == "__main__":main()
Enter the following command to run this script:
python main.pyThe initial cell configuration is set by this statement:
cells = [(2, 4), (2, 5), (1, 5), (1, 6), (3, 5)]You can try another configuration by uncommenting the prior cells array, and then comment out this one. I did that very action and then ran the script. I observed an unusual display, which I will not describe. I will leave it for you to discover.
I want to caution that what I am about to describe can become quite addictive. It is testing the consequences of new initial starting patterns. There are more than a few AI researchers who have dedicated their careers to the study of cellular automata, which includes researching the fascinating evolving patterns from the Game of Life.
Figure 10-11 illustrates some starting configurations that you may wish to try. The companion cell configuration array values are shown next to each of the patterns.
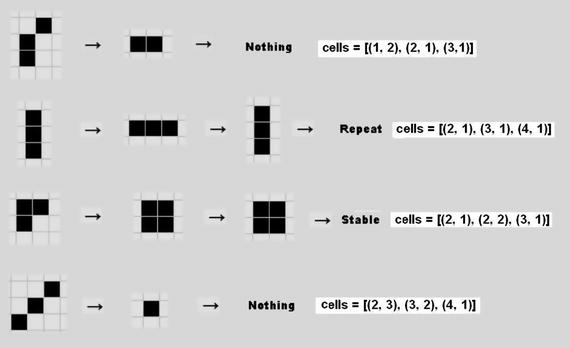
Figure 10-11. Example Game of Life starting patterns
Two of the patterns immediately disappear (die), one goes into a bi-stable state, and the fourth pattern enters a stable state. I tested each pattern and confirmed it acted as shown.
Figure 10-12 shows other initial patterns that you can experiment with to see how they evolve according to the rules set. The companion cell array values are shown next to each pattern.
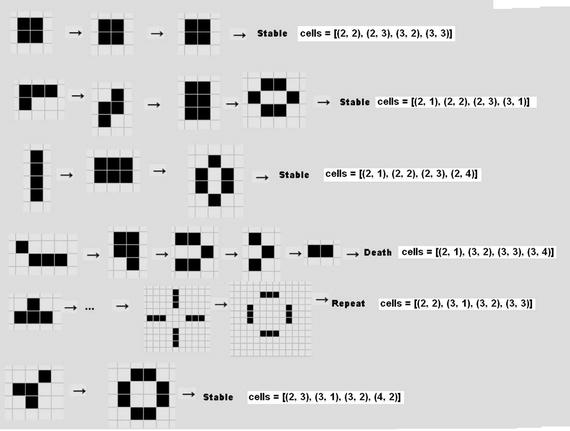
Figure 10-12. Additional starting patterns
There are a series of patterns that are dynamic, which means that they constantly move across the grid and repeat their patterns. Figure 10-13 shows the glider that moves around the grid and repeats its pattern every fourth generation.
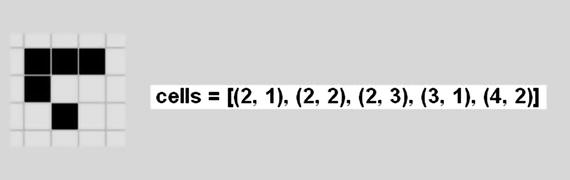
Figure 10-13. Glider pattern
A similar dynamic pattern is the lightweight spaceship, which is shown in Figure 10-14. It moves across the grid too.

Figure 10-14. Lightweight spaceship pattern
Conway discovered several patterns that took many generations to finally evolve and become both predictable and periodic. Incidentally, he made these discoveries without the aid of a computer. He called these patterns Methuselahs, after a man who was described in the Hebrew Bible to have lived to the age of 969 years. The first of these long-lived patterns is named F-pentomino, which is shown in Figure 10-15. It becomes stable after 1101 generations.

Figure 10-15. F-pentomino pattern
The Acorn pattern shown in Figure 10-16 is another example of a Methuselah that becomes stable and predictable after 5206 generations.

Figure 10-16. Acorn pattern
Readers who wish to experiment with more patterns can go to Alan Hensel’s webpage at radicaleye.com/lifepage/picgloss/picgloss.html, where he has compiled a fairly large list of other common patterns.
This completes the initial foray into cellular automata using Conway’s Game of Life as the tool. You should now be empowered to further experiment with this tool to gain more experience and confidence in this powerful AI topic. I also highly recommend Dr. Stephen Wolfram’s book A New Kind of Science (Wolfram Media, 2002), in which he examines the entire field of cellular automata using the Mathematica application, which he created. Incidentally, the Mathematica application is now freely provided with the latest Raspian distributions available from raspberrypi.org.
Summary
This chapter was concerned with evolutionary computing. I began the chapter with a story relating how evolution and mutation were integral parts of EC.
The first demonstration showed how evolutionary programming could be used to find solutions to fairly simple problems using both evolution and mutation techniques. The solution was first calculated manually and then automatically by a Python script.
The second demonstration introduced the EC subtopic of genetic algorithms and genetic programming. I used a Python version of Conway’s Game of Life as the means to explain and demonstrate these concepts. This section also introduced the concept of cellular automata, which is central to the game.
There were two game versions shown: one that used genetic evolution and mutation, and another that was more deterministic in that you could specify the starting patterns. The latter version was further used to examine a variety of cellular patterns that generated some unusual behaviors.
A Sense HAT accessory board was used with a Raspberry Pi 3 to display the Game of Life simulations.
