This is ordinary linear regression!
Although xi is a scalar here, the method generalizes to a vector xi. Categorical variables can be encoded into a subset of the dimensions of x using the so-called “one-hot” method, placing a 1 in the dimension corresponding to the category label and 0s in all other dimensions allocated to the variable. If all input variables are categorical, this corresponds to the classic analysis of variance (ANOVA) method.
Using Priors on Parameters
Placing a Gaussian prior on the parameters w leads to the method of ridge regression (Section 7.2)— also called “weight decay.” Consider a regression that uses a D-dimensional vector x to make predictions. The regression’s bias term can be represented by defining the first dimension of x as the constant 1 for every example. Defining ![]() and using the usual
and using the usual ![]() notation for scalar Gaussians, the underlying probability model is
notation for scalar Gaussians, the underlying probability model is
where τ is the hyperparameter specifying the prior. Setting ![]() , it is possible to show that maximum a posteriori parameter estimation based on the log conditional likelihood is equivalent to minimizing the squared error loss function
, it is possible to show that maximum a posteriori parameter estimation based on the log conditional likelihood is equivalent to minimizing the squared error loss function
(9.4)
which includes an L2-based regularization term given by ![]() . (“Regularization” is another term for overfitting avoidance.)
. (“Regularization” is another term for overfitting avoidance.)
Using a Laplace prior for the distribution over weights and taking the log of the likelihood function yields an L1-based regularization term ![]() . To see why, note that the Laplace distribution has the form
. To see why, note that the Laplace distribution has the form
where ![]() and b are parameters. Modeling the prior probability for each weight by a Laplace distribution with
and b are parameters. Modeling the prior probability for each weight by a Laplace distribution with ![]() yields
yields
Since the Laplace distribution places more probability at zero than the Gaussian distribution does, it can provide both regularization and variable selection in regression problems. This technique has been popularized as a regression approach known as the LASSO, “Least Absolute Shrinkage and Selection Operator.”
An alternative approach known as the “elastic net” combines L1 and L2 regularization techniques using
This corresponds to a prior distribution that consist of the product of a Gaussian and a Laplacian distribution. The result leads to convex optimization problems—i.e., problems where any local minimum must be a global minimum—if the loss is convex, which applies to models like logistic or linear regression.
Matrix vector formulations of linear and polynomial regression
This section formulates linear regression using matrix operations. Observe that the loss in (9.4)—without the penalty term—can be written

where the vector y is just the individual yi’s stacked up, and w is the vector of parameters (or weights) for the model. Taking the partial derivative with respect to w and setting the result to zero yields a closed form expression for the parameters:
(9.5)
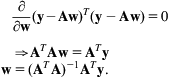 (9.5)
(9.5)
These are famous equations. ![]() is known as the normal equations, and the quantity
is known as the normal equations, and the quantity ![]() is known as the pseudoinverse. Note that
is known as the pseudoinverse. Note that ![]() is not always invertible, but this problem can be addressed using regularization.
is not always invertible, but this problem can be addressed using regularization.
For ridge regression, a prior is added to make the objective function
(9.6)
for a suitably defined matrix A with vectors xi in the rows, and ![]() as defined above. Again, setting the partial derivative of F(w) to zero yields a closed form solution:
as defined above. Again, setting the partial derivative of F(w) to zero yields a closed form solution:

This modification to the pseudoinverse equation allows solutions to be found that would otherwise not exist, often using a very small ![]() . It is often presented as a regularization method that avoids numerical instability, but the analysis in terms of a prior on parameters gives more insight. For example, it may be appropriate to use Gaussian priors of different strengths for different terms. In fact, it is common practice to impose no penalty at all on the bias weight. This could be implemented by replacing the
. It is often presented as a regularization method that avoids numerical instability, but the analysis in terms of a prior on parameters gives more insight. For example, it may be appropriate to use Gaussian priors of different strengths for different terms. In fact, it is common practice to impose no penalty at all on the bias weight. This could be implemented by replacing the ![]() term in (9.6) by
term in (9.6) by ![]() , where D is a diagonal matrix containing the λi’s to be used for each weight, transforming the solution into
, where D is a diagonal matrix containing the λi’s to be used for each weight, transforming the solution into ![]() . While the above expression for the partial derivative is fairly simple, care must still be taken during implementation to avoid numerically instable results.
. While the above expression for the partial derivative is fairly simple, care must still be taken during implementation to avoid numerically instable results.
The linear regression model can be transformed into a nonlinear polynomial model. Although the polynomial regression model yields nonlinear predictions, the estimation problem is linear in the parameters. To see this, express the problem in matrix form, using a suitably defined matrix A to encode the polynomial’s higher order terms and a vector c to encode the coefficients, including those used for the higher order terms:

Eq. (9.5) can be used to solve for the parameters in closed form.
This trick of transforming a linear prediction method into a nonlinear one while keeping the underlying estimation problem linear can be generalized. The general approach goes by the name of basis function expansion. For polynomial regression, the polynomial basis given by the rows of the matrix A (the powers of x) is used. However, any nonlinear function of the inputs ![]() could be used to define models of the form
could be used to define models of the form
which also give closed form solutions for a linear parameter estimation problem. We will return to this when discussing kernelizing probabilistic models below.
Multiclass Logistic Regression
Binary logistic regression was introduced in Section 4.6. Consider now a multiclass classification problem where class values are encoded as instances of the random variable ![]() , and, as before, feature vectors are instances of a variable x. Assume there is no significance to the order of the classes.
, and, as before, feature vectors are instances of a variable x. Assume there is no significance to the order of the classes.
A simple linear probabilistic classifier can be created using this parametric form:
(9.7)
which employs K feature functions fk(y,x) and K weights, wk as the parameters of the model. This is one way of formulating multinomial logistic regression. The feature functions could encode complex features extracted from the input vector x. To perform learning using maximum conditional likelihood and observations that are instances of y and x, ![]() , write the objective function as
, write the objective function as
Unfortunately there is no closed form solution for the parameter values that maximize this conditional likelihood, and optimization is usually performed using a gradient based procedure. On taking the partial derivative with respect to one of the weights of the log conditional probability for just one observation we have

The derivative breaks apart into two terms. The first is easy because all terms involving weights where ![]() are 0, leaving the sum with just one term. The second, while seemingly daunting, has a derivative that yields an intuitive and interpretable result:
are 0, leaving the sum with just one term. The second, while seemingly daunting, has a derivative that yields an intuitive and interpretable result:

which corresponds to the expectation ![]() of the feature function
of the feature function ![]() under the probability distribution
under the probability distribution ![]() given by the model with the current parameter settings. Write the feature functions as a function
given by the model with the current parameter settings. Write the feature functions as a function ![]() in vector form. Then for a vector of weights w, the partial derivative of the conditional log-likelihood for the entire dataset with respect to w is
in vector form. Then for a vector of weights w, the partial derivative of the conditional log-likelihood for the entire dataset with respect to w is
This consists of the sum of the differences between the observed feature vector for a given example and the expected value of the feature vector under the current model settings. If the model were perfect, classifying each example correctly with probability 1, the partial derivative would be zero. Intuitively, the learning procedure adjusts the model parameters so as to produce predictions that are closer to the observed data.
Matrix vector formulation of multiclass logistic regression
The model in (9.7) for a multiclass linear probabilistic classifier using vectors wc for the weights associated with each class can be written
where y is an index, the weights are encoded into a vector of length K, and the features x have been re-defined as the result of evaluating the feature functions fk(y,x) in such a way that there is no difference between the features given by fk(y=i,x) and fk(y=j,x). This form is widely used for the last layer in neural network models, where it is referred to as the softmax function.
The information concerning class labels can be encoded into a multinomial vector y, which is all 0s except for a single 1 in the dimension that represents the correct class label—e.g., ![]() for the second class. The weights form a matrix
for the second class. The weights form a matrix ![]() , and the biases form a vector
, and the biases form a vector ![]() . Then the model yields vectors of probabilities
. Then the model yields vectors of probabilities

where the denominator sums over each possible label ![]() , which is
, which is ![]() . Re-defining x as x=[xT 1]T and the parameters as a matrix of the form
. Re-defining x as x=[xT 1]T and the parameters as a matrix of the form

the conditional model can be written in this compact matrix-vector form:
Then the gradient of the log-conditional-likelihood with respect to the parameter matrix θ can be expressed as

where the notation ![]() denotes the expectation of the random vector y under
denotes the expectation of the random vector y under ![]() , the vector of conditional probabilities that the model yields for the observed input
, the vector of conditional probabilities that the model yields for the observed input ![]() under the current parameter settings. The first term corresponds to a matrix that is computed just once, when the procedure commences. The second corresponds to a matrix that approaches the first term more closely as the model learns to make predictions that match the observed data.
under the current parameter settings. The first term corresponds to a matrix that is computed just once, when the procedure commences. The second corresponds to a matrix that approaches the first term more closely as the model learns to make predictions that match the observed data.
Formulating these equations as vector and matrix operations allows highly optimized numerical libraries to be applied. Fast libraries for vector and matrix operations are key facilitators of big data techniques. In particular, state-of-the-art methods for deep learning with large datasets rely heavily on the dramatic performance improvements enabled by GPUs.
Note that we have not taken account of the fact that the final prediction need only yield probabilities for N–1 of the N classes, the remaining one being inferred from the fact that they must all sum to 1. Multinomial logistic regression models can be formulated that exploit this fact and involve fewer parameters.
Priors on parameters, and the regularized loss function
Logistic regression is frequently performed with some regularizer or prior on the parameters to combat overfitting. From a probabilistic perspective, this means that the conditional probability for the set of all labels Y given the set of all input vectors X can be rewritten
where ![]() is a prior distribution for the parameters. Given observed data
is a prior distribution for the parameters. Given observed data ![]() , finding the value of
, finding the value of ![]() that maximizes this expression is an instance of maximum a posteriori parameter estimation with a conditional probability model. The goal, then, is to minimize
that maximizes this expression is an instance of maximum a posteriori parameter estimation with a conditional probability model. The goal, then, is to minimize
The first term is the negative log-likelihood, corresponding to the loss function, and the second is the negative log of the prior for the parameters, also known as the “regularization” term. L2 regularization is often used for the weights in a logistic regression model. A prior could be applied to the bias too; but in practice it is often better not to do this (or, equivalently, to use a uniform distribution as the prior).
So-called “L2 regularization” is based on the L2 norm, which is just the Euclidean distance ![]() . If a Gaussian distribution with zero mean and a common variance for each weight is used as the prior for the weights, the corresponding regularization term is the squared L2 distance weighted by
. If a Gaussian distribution with zero mean and a common variance for each weight is used as the prior for the weights, the corresponding regularization term is the squared L2 distance weighted by ![]() , plus a constant:
, plus a constant:
The constant can be ignored during the optimization and is often omitted from the regularized loss function. However, using ![]() or
or ![]() as the regularization term gives a more direct correspondence with the
as the regularization term gives a more direct correspondence with the ![]() parameter of the Gaussian distribution.
parameter of the Gaussian distribution.
Solving a regularized multiclass logistic regression with M weight parameters corresponds to finding the weights and biases that minimize
the regularization term will encourage the weights to be small. In the context of multilayer perceptrons, this sort of regularization is known as weight decay; in statistics it is known as ridge regression.
In regression, the elastic net regularization introduced earlier combines L1 and L2 regularization using ![]() , which corresponds to a prior on weights consisting of the product of Laplacian and Gaussian distributions. In terms of a matrix representation, given a weight matrix
, which corresponds to a prior on weights consisting of the product of Laplacian and Gaussian distributions. In terms of a matrix representation, given a weight matrix ![]() with row and column entries given by
with row and column entries given by ![]() , this can be written
, this can be written

where we have set the derivative of the L1 prior to zero at zero despite the fact that it is technically undefined at this point. This is common practice, since the goal of this type of regularization is to induce sparsity: once a weight is zero the gradient will be zero. Notice that regularization has not been applied to the bias terms. This regularizer leads to convex optimization problems if the loss is convex, which is the case for logistic regression and linear regression.
Gradient Descent and Second-Order Methods
By formulating maximum likelihood learning with a prior on parameters as the problem of minimizing the negative log probability, gradient descent can be used to optimize the model’s parameters. Given a conditional probability model ![]() with parameter vector
with parameter vector ![]() and data
and data ![]() , along with a prior on parameters given by
, along with a prior on parameters given by ![]() , with hyperparameter
, with hyperparameter ![]() , the gradient descent procedure with learning rate
, the gradient descent procedure with learning rate ![]() is:
is:

Convergence is usually determined by monitoring the change in the loss or the parameters and terminating when one of them stabilizes. Appendix A.1 shows how a Taylor series expansion can be used to interpret and justify the learning rate parameter ![]() .
.
Alternatively, gradient descent can be based on the second derivative by computing the Hessian matrix H at each iteration and replacing the above update by
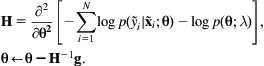
Generalized Linear Models
Linear regression and logistic regression are special cases of a family of conditional probability models known in statistics as “generalized linear models,” which were proposed to unify and generalize linear and logistic regression. In this formulation, linear models may be related to a response variable using distributions other than the Gaussian distribution used for linear regression. Generalized linear models can be created for any distribution in the exponential family (Appendix A.2 introduces exponential-family distributions).
In this context, the data can be thought of in terms of response variables yi and explanatory variables organized as p-dimensional vectors xi, i=1,…, n. Response variables can be expressed in different ways, ranging from binary to categorical or ordinal data. A model is then defined where the expected value ![]() of the distribution used for the response variable consists of an initial linear prediction
of the distribution used for the response variable consists of an initial linear prediction ![]() using a parameter vector
using a parameter vector ![]() , which is then subjected to a smooth, invertible, and potentially nonlinear transformation using the mean function g−1:
, which is then subjected to a smooth, invertible, and potentially nonlinear transformation using the mean function g−1:
The mean function is the inverse of the link function g. In generalized linear modeling the entire set of explanatory variables for all the observations is arranged as an ![]() matrix X, so that a vector of linear predictions for the entire data set is
matrix X, so that a vector of linear predictions for the entire data set is ![]() . The variance of the underlying distribution can also be modeled; typically as a function of the mean. Different distributions, link functions, and corresponding mean functions give a great deal of flexibility in defining probabilistic models; Table 9.2 shows some examples.
. The variance of the underlying distribution can also be modeled; typically as a function of the mean. Different distributions, link functions, and corresponding mean functions give a great deal of flexibility in defining probabilistic models; Table 9.2 shows some examples.
Table 9.2
Link Functions, Mean Functions, and Distributions Used in Generalized Linear Models
| Link Name | Link Function
|
Mean Function
|
Typical Distribution |
| Identity | Gaussian | ||
| Inverse | Exponential | ||
| Log | Poisson | ||
| Log-log | Bernoulli | ||
| Logit | Bernoulli | ||
| Probit | Bernoulli |

The multiclass extension of logistic regression discussed above is another example of a generalized linear model that uses the multinomial distribution for the response variable y. Because this model is defined in terms of probability distributions, the parameters can be estimated using maximum likelihood techniques.
Since these models are fairly simple, the coefficients ![]() are interpretable. Applied statisticians are often interested not only in the estimated values but in other information such as the standard error of the estimates and statistical significance tests.
are interpretable. Applied statisticians are often interested not only in the estimated values but in other information such as the standard error of the estimates and statistical significance tests.
Making Predictions for Ordered Classes
In many situations class values are categorical but possess a natural ordering. To deal with ordinal class attributes, the class probabilities can be expressed in terms of cumulative distributions, which are then modeled to construct an underlying probability distribution function for each class. To define a model with M ordinal categories, M–1 cumulative probability models of the form ![]() are used for the random variable Yi that represents the category of a given instance i. Models for
are used for the random variable Yi that represents the category of a given instance i. Models for ![]() are then obtained using differences between the cumulative distribution models. Here we will use complementary cumulative probabilities, known as survival functions, of the form
are then obtained using differences between the cumulative distribution models. Here we will use complementary cumulative probabilities, known as survival functions, of the form ![]() , because this sometimes simplifies the interpretation of parameters. Then the class probabilities are obtained by:
, because this sometimes simplifies the interpretation of parameters. Then the class probabilities are obtained by:
The generalized linear models discussed above can be further generalized to ordinal categorical data. In fact, the general approach can be applied to various model classes by modeling complementary cumulative probabilities with a smooth and invertible link function that transforms them nonlinearly and equates the result to a linear predictor. For binary predictions, the models often take this form:
where w is a vector of weights, xi are vectors of features, and ![]() represent the probability that example i is greater than the discretized, ordinal category j. Such models are called “proportional odds” models, or “ordered logit” models. The model above uses a different bias for each inequality, but the same set of weights. This guarantees a consistent set of probabilities.
represent the probability that example i is greater than the discretized, ordinal category j. Such models are called “proportional odds” models, or “ordered logit” models. The model above uses a different bias for each inequality, but the same set of weights. This guarantees a consistent set of probabilities.
Conditional Probabilistic Models Using Kernels
Linear models can be transformed into nonlinear ones by applying the “kernel trick” mentioned in Section 7.2 under kernel regression, or, alternatively, through basis expansion as mentioned in the section above on matrix formulations of linear and polynomial regression. This can be applied to both kernel regression and kernel logistic regression.
Suppose the features x are replaced by the vector k(x) whose elements are determined using a kernel function k(x,xj) for every training example (or for some subset of training vectors):

A “1” has been appended to this vector to implement the bias term in the parameter matrix. A kernel regression is then
For classification, an analogous kernel logistic regression is
Since every example in the training set can have a different kernel vector, it is necessary to compute a kernel matrix K with entries given by Kij=k(xi,xj). For large datasets this operation can be time- and memory-intensive.
Support vector machines have a similar form, although they are not formulated probabilistically, and multiclass support vector machines are not as easily formulated as multiclass kernel logistic regression. Because they use an underlying hinge loss and weight regularization term, support vector machines often assign zero weights to many of the terms; they also have the appealing feature of placing nonzero weights only on vectors that reside at the boundary of the decision surface. In many applications this results in a significant reduction in the number of kernel evaluations needed at test time. Neither kernel logistic regression nor kernel ridge regression produce such sparse solutions, even when a Gaussian or (squared) L2 regularization is used: in general there is a nonzero weight for every exemplar. However, kernel logistic regression can outperform support vector machines, and several techniques have been proposed to make these methods sparse. They are mentioned in the Further Reading section at the end of the chapter.
9.8 Sequential and Temporal Models
Consider the task of creating a probability model for a sequence of observations. If they correspond to words, random variables could be defined with as many states as there are words in the vocabulary. If they are continuous, some parametric form is needed to create suitable continuous distributions.
Markov Models and N-gram Methods
One simple and effective probabilistic model for discrete sequential data is known as a “Markov model.” A first-order Markov model assumes that each symbol in a sequence can be predicted using its conditional probability given the preceding symbol. (For the very first symbol, an unconditional probability is used.) Given observation variables O={O1,…,OT}, this can be written
Usually, every conditional probability used in such models is the same. The corresponding Bayesian networks consist simply of a linear chain of variables with directed edges between each successive pair; Fig. 9.18A shows an example. The method generalizes naturally to second-order models (Fig. 9.18B), and to higher orders. N-gram models correspond to the use of an (N–1)-th order Markov model. For example, first-order models involve the use of 2-grams (or “bigrams”); third-order models use trigrams, and zero-th order models correspond to single observations or “unigrams.” Such models are widely used for modeling biological sequences such as DNA, as well as for text mining and computational linguistics applications.

All probabilistic models raise the issue of what to do when there is no data for certain configurations of variables. Zero-valued parameters cause problems, as we saw in Section 4.2. This is particularly severe with high-order Markov models, and smoothing techniques—such as Laplace or Dirichlet smoothing, which can be derived from a Bayesian analysis—become critical. See the Further Reading section for some pointers to some specialized methods for smoothing n-grams.
Large n-gram models are extremely useful for applications ranging from machine translation and speech recognition to spelling correction and information extraction. Indeed, Google has made available English language word counts for a billion five-word sequences that appear at least 40 times in a trillion-word corpus After discarding words that appear less than 200 times, there remain 13 million unique words (unigrams), 300 million bigrams; and around a billion each of trigrams, four-grams, and five-grams.
Hidden Markov Models
Hidden Markov models have been widely used for pattern recognition since at least the 1980s. Until recently most of the major speech recognition systems have consisted of large Gaussian mixture models combined with hidden Markov models. Many problems in biological sequence analysis can also be formulated in terms of hidden Markov models, with various extensions and generalizations.
A hidden Markov model is a joint probability model of a set of discrete observed variables O={O1,…,OT} and discrete hidden variables H={H1,…,HT} for T observations that factors the joint distribution as follows:
Each Ot is a discrete random variable with N possible values, and each Ht is a discrete random variable with M possible values. Fig. 9.18C illustrates a hidden Markov model as a type of Bayesian network that is known as a “dynamic” Bayesian network because variables are replicated dynamically over the appropriate number of time steps. They are an obvious extension of first-order Markov models, and it is common to use “time-homogeneous” models where the transition matrix P(Ht+1|Ht) is the same at each time step. Define A to be a transition matrix whose elements encode P(Ht+1=j|Ht=i), and B to be an emission matrix B whose elements bij correspond to P(Ot=j|Ht=i). For the special t=1 the initial state probability distribution is encoded in a vector π with elements πi=P(Ht=i). The complete set of parameters is ![]() , a set containing two matrices and one vector. We write a particular observation sequence as a set of observations
, a set containing two matrices and one vector. We write a particular observation sequence as a set of observations ![]() .
.
Hidden Markov models pose three key problems:
1. Compute ![]() , the probability of a sequence under the model with parameters
, the probability of a sequence under the model with parameters ![]() .
.
2. Find the most probable explanation—the best sequence of states ![]() that explains an observation.
that explains an observation.
3. Find the best parameters ![]() for the model given a data set of observed sequences.
for the model given a data set of observed sequences.
The first problem can be solved using the sum-product algorithm, the second using the max-product algorithm, and the third using the EM algorithm for which the required expectations are computed using the sum-product algorithm. If there is labeled data for the sequences of hidden variables that correspond to observed sequences, the required conditional probability distributions can be computed from the corresponding counts in the same way as we did with Bayesian networks.
The only difference between the parameter estimation task when using an hidden Markov model viewed as a dynamic Bayesian network and the updates used for learning the conditional probability tables in a Bayesian network is that one can average over the statistics obtained at each time step, because the same emission and transition matrices are used at each step. The basic hidden Markov model formulation serves as a point of reference when coupling more complex probabilistic models over time using more general dynamic Bayesian network models.
Conditional Random Fields
Hidden Markov models are not the only models that are useful when working with sequence data. Conditional random fields are a statistical modeling technique that takes context into account, and are often structured as linear chains. They are widely used for sequence processing tasks in data mining, but are also popular in image processing and computer vision. Fig. 9.19 illustrates the problem of extracting locations and dates of meetings from email text, which can be addressed using chain-structured conditional random fields. A major advantage is that these models can be given arbitrarily complex features of the input sequence to be used for making predictions—e.g., matching words to lists of organizations and processing variations of known abbreviations.
Inference for chain-structured conditional random fields can be performed efficiently using the sum- and max-product algorithms discussed above. The sum-product algorithm can be used to compute the expected gradients needed to learn them, and the max-product algorithm serves to label new sequences—with tags such as “where” indicating the location or room and “time” labels associated with a meeting.
We begin with a general definition, and then focus on the simpler case of linear conditional random fields. Fig. 9.20D shows a chain-structured conditional random field, illustrated as a factor graph, which can be contrasted with the Markov random field depicted in Fig. 9.20B and the hidden Markov model in Fig. 9.20C, also shown as a factor graph. Circles are not used to represent the observed variables in Fig. 9.20D because the underlying conditional random field model does not explicitly encode these as random variables in the graph.

From Markov random fields to conditional random fields
Whereas both Bayesian networks and Markov random fields define a joint probability model for data, conditional random fields (also known as “structured prediction” techniques) define a joint conditional distribution for multiple predictions. For a set X of random variables, we have already seen how a Markov random field factorizes the joint distribution for X using an exponentiated energy function F(X):

where the sum is over all states of all variables in X. Conditional random fields condition on some observations, yielding a conditional distribution:

where the sum is over all states of all variables in Y. Both Markov and conditional random fields can be defined for general model structures, but the energy functions usually include just one or two variables—unary and pairwise potentials. Conceptually, to create a conditional random field for P(Y|X) based on U unary and V pairwise functions of variables in Y, the energy function takes the form
Such energy functions can be transformed into so-called potential functions by negation, exponentiation, and normalization. The conditional probability model takes this form:
(9.8)
 (9.8)
(9.8)Lattice-structured models like this are used in image processing applications, as mentioned earlier. Processing sequences with chain-structured conditional random fields is even simpler. Note that logistic regression could be thought of as a simple conditional random field, one that arises naturally from a conditional version of a Markov random field with a factorization shown in Fig. 9.15B.
Linear chain conditional random fields
Consider observations ![]() of a sequence of discrete random variables
of a sequence of discrete random variables ![]() , using integers to encode the relevant states, and an observed input sequence
, using integers to encode the relevant states, and an observed input sequence ![]() , which can be of any data type. A conditional random field defines the conditional probability
, which can be of any data type. A conditional random field defines the conditional probability ![]() of a label sequence given an input sequence. This contrasts with hidden Markov models, which treat both Y and X as random variables and defines a joint probability model for P(Y,X). Note that X above was intentionally not defined as a sequence of random variables because we will define the conditional distribution of Y given X and need not have an explicit model for P(X). Of course, an implicit model for P(X) could be defined as the empirical distribution of the data, or the distribution produced by placing a Dirac or Kronecker delta function on each observation and normalizing by the number of examples. Not defining X as a sequence leaves open what type of variables these are, and whether it is just a set of variables or a set of formally defined random variables. Fig. 9.20D uses shaded rectangles to emphasize this point. In this chain model, for a given sequence of length N, Eq. (9.8) could be re-written
of a label sequence given an input sequence. This contrasts with hidden Markov models, which treat both Y and X as random variables and defines a joint probability model for P(Y,X). Note that X above was intentionally not defined as a sequence of random variables because we will define the conditional distribution of Y given X and need not have an explicit model for P(X). Of course, an implicit model for P(X) could be defined as the empirical distribution of the data, or the distribution produced by placing a Dirac or Kronecker delta function on each observation and normalizing by the number of examples. Not defining X as a sequence leaves open what type of variables these are, and whether it is just a set of variables or a set of formally defined random variables. Fig. 9.20D uses shaded rectangles to emphasize this point. In this chain model, for a given sequence of length N, Eq. (9.8) could be re-written
Focusing on a linear chain structure reveals some analogies with the emission and transition matrices of hidden Markov models. There are two types of features: a set of J single variable (state) features ![]() that are a function of a single yi, and which are computed for each yi in the sequence i=1,…, N; and a set of K pairwise (transition) features
that are a function of a single yi, and which are computed for each yi in the sequence i=1,…, N; and a set of K pairwise (transition) features ![]() for i>1. Each type has associated unary weights
for i>1. Each type has associated unary weights ![]() and pairwise weights
and pairwise weights ![]() . Note that these features can be a function of the entire observed sequence
. Note that these features can be a function of the entire observed sequence ![]() , or of some subset. This global dependence on the input is a major advantage of conditional random fields over hidden Markov models. The analog of the Markov model transition matrix are the per-position pairwise potential functions: these can be written as a set of matrices that depend upon the observation sequence and consist of a sum over the product of pairwise weights
, or of some subset. This global dependence on the input is a major advantage of conditional random fields over hidden Markov models. The analog of the Markov model transition matrix are the per-position pairwise potential functions: these can be written as a set of matrices that depend upon the observation sequence and consist of a sum over the product of pairwise weights ![]() and pairwise features
and pairwise features
The unary potential functions play a similar role to the terms arising from the hidden Markov model emission matrices, and can be written as the following set of vectors that involve exponentiated weighted combinations of unary features
Sometimes, rather than keeping the unary and pairwise features and their parameters separate, it is useful to work with all features for a given position i. To do this, define ![]() as a length-L vector containing all single-variable and pairwise features, and define the global feature vector to be the sum over each of these position dependent feature vectors:
as a length-L vector containing all single-variable and pairwise features, and define the global feature vector to be the sum over each of these position dependent feature vectors:
Now a conditional random field can be written in a particularly compact form
If we turn to the problem of learning based on maximizing the conditional likelihood for this model, an analogy can be drawn to the simpler case of logistic regression. The gradient of the log-likelihood of a conditional random field for a set of M input sequences ![]() and corresponding output sequences
and corresponding output sequences ![]() , is
, is
Here, Em[.] is an expectation taken with respect to ![]() . It is standard practice to include a Gaussian prior or L2 regularization on parameters by adding a regularization term to this expression. Unlike logistic regression, this expectation involves the joint distribution of the label sequence and not just the distribution for a single label variable. However, in chain-structured graphs it can be computed efficiently and exactly using the sum-product algorithm.
. It is standard practice to include a Gaussian prior or L2 regularization on parameters by adding a regularization term to this expression. Unlike logistic regression, this expectation involves the joint distribution of the label sequence and not just the distribution for a single label variable. However, in chain-structured graphs it can be computed efficiently and exactly using the sum-product algorithm.
Learning for chain-structured conditional random fields
To compute the gradient for a linear chain conditional random field, it is useful to expose each parameter in the L2 regularized log-likelihood in scalar form:
where σ is the regularization parameter. Taking the derivative with respect to a single parameter and training example, the contribution to the gradient of each example is
This is simply the difference between the observed occurrence of the feature and its expectation under the current prediction of the model, taken with respect to ![]() , minus the partial derivative of the regularization term. Similar terms arise for unary functions, but the expectation is taken with respect to
, minus the partial derivative of the regularization term. Similar terms arise for unary functions, but the expectation is taken with respect to ![]() . These distributions are the single- and pairwise-variable marginal conditional distributions, and can be computed efficiently using the sum-product algorithm.
. These distributions are the single- and pairwise-variable marginal conditional distributions, and can be computed efficiently using the sum-product algorithm.
Using conditional random fields for text mining
The text information extraction scenario in Fig. 9.19 is just one example of applying data mining to extract information from natural language. Such information might be other named entities such as locations, personal names, organizations, money, percentages, dates, and times; or fields in a seminar announcement such as speaker name, seminar room, start time, and end time. In such tasks, input features often consist of current, previous, and next word; character n-grams; part-of-speech tag sequences; the presence of certain key words in windows to the left or right of the current position. Other features can be defined using lists of known words, such as first and last names and honorifics; locations and organizations. Features such as capitalization and alphanumeric characters can be defined using regular expressions and integrated into an underlying probabilistic model based on conditional random fields.
9.9 Further Reading and Bibliographic Notes
The field of probabilistic machine learning and data mining is enormous: it essentially subsumes all classical and modern statistical techniques. This chapter has focused on foundational concepts and some widely used probabilistic techniques in data mining and machine learning. Excellent books that focus on statistical and probabilistic methods include Hastie, Tibshirani, and Friedman (2009), and Murphy (2012). Koller and Friedman (2009)’s excellent book specializes in advanced techniques and principles of probabilistic graphical models.
The K2 algorithm for learning Bayesian networks was introduced by Cooper and Herskovits (1992). Bayesian scoring metrics are covered by Heckerman et al. (1995). Friedman, Geiger, and Goldszmidt (1997) introduced the tree augmented Naïve Bayes algorithm, and also describe multinets. Grossman and Domingos (2004) show how to use the conditional likelihood for scoring networks. Guo and Greiner (2004) present an extensive comparison of scoring metrics for Bayesian network classifiers. Bouckaert (1995) describes averaging over subnetworks. AODEs are described by Webb, Boughton, and Wang (2005), and AnDEs by Webb et al. (2012). AD trees were introduced and analyzed by Moore and Lee (1998)—the same Andrew Moore whose work on kD-trees and ball trees was mentioned in Section 4.10. Komarek and Moore (2000) introduce AD trees for incremental learning that are also more efficient for data sets with many attributes.
The AutoClass program is described by Cheeseman and Stutz (1995). Two implementations have been produced: the original research implementation, written in LISP, and a follow-up public implementation in C that is 10 or 20 times faster but somewhat more restricted—e.g., only the normal distribution model is implemented for numeric attributes. DensiTrees were developed by Bouckaert (2010).
Kernel density estimation is an effective and conceptually simple probabilistic model. Epanechnikov (1969) showed the optimality of the Epanechnikov kernel under the mean-squared error metric. Jones, Marron, and Sheather (1996) recommends using a so-called “plug-in” estimate to select the kernel bandwidth. Duda and Hart (1973) and Bishop (2006) show theoretically that kernel density estimation converges to the true distribution as the amount of data grows.
The EM algorithm, which originates in the work of Dempster, Laird, and Rubin (1977), is the key to learning in hidden or latent variable models. The modern variational view provides a solid theoretical justification for the use of approximate posterior distributions, as discussed in Appendix A.2 and in Bishop (2006). This perspective originated in the 1990s with work by Neal and Hinton (1998), Jordan, Ghahramani, Jaakkola, and Saul (1998), and others. Salakhutdinov, Roweis and Ghahramani (2003) explore the EM approach and compare it with the expected gradient, including the more sophisticated expected conjugate gradient based optimization.
Markov chain Monte Carlo methods are popular in Bayesian statistical modeling; see, e.g., Gilks (2005). Geman and Geman (1984) first described the Gibbs sampling procedure, naming it after the physicist Josiah Gibbs because of the analogy between sampling, the underlying functional forms of random fields and statistical physics. Hastings’ (1970) generalization of the Metropolis, Rosenbluth, Rosenbluth, Teller, and Teller (1953) algorithm has been influential in laying the foundation for present-day methods. The iterated conditional modes approach for finding an approximate most probable explanation was proposed by Besag (1986).
Plate notation has been widely used in artificial intelligence (Buntine, 1994), machine learning (Blei, Ng, & Jordan, 2003) and computational statistics (Lunn, Thomas, Best, & Spiegelhalter, 2000) to define complex probabilistic graphical models, and forms the basis of the BUGS (Bayesian inference Using Gibbs Sampling) software project (Lunn et al., 2000). Our presentation of factor graphs and the sum-product algorithm follows their origins in Kschischang, Frey, and Loeliger (2001) and Frey (1998). The sum- and max-product algorithms only apply to trees. However, Bayesian networks and other models that contain cycles can be manipulated into a structure known as a junction tree by clustering variables, and Lauritzen and Spiegelhalter (1988)’s junction tree algorithm permits exact inference. Ripley (1996) covers the junction tree algorithm, with practical examples; Huang and Darwiche (1996)’s procedural guide is an excellent resource for those who need to implement the algorithm. Probability propagation in a junction tree yields exact results, but is sometimes infeasible because the clusters become too large—in which case one must resort to sampling or variational methods.
Roweis (1998) gives an early EM formulation for PPCA: he examines the zero input noise case and provides the elegant mathematics for the simplified EM algorithm presented above. Tipping and Bishop (1999a, 1999b) give further analysis, and show that after optimizing the model and learning the variance of the observation noise the columns of the matrix W are scaled and rotated principal eigenvectors of the covariance matrix of the data. The probabilistic formulation of principal component analysis opens the door to probabilistic formulations of further generalizations, such as mixtures of principal component analyzers (Dony & Haykin, 1995; Tipping & Bishop, 1999a, 1999b) and mixtures of factor analyzers (Ghahramani & Hinton, 1996). Of particular utility is the ability of PPCA and factor analysis to easily deal with missing data: providing the data is missing at random one can marginalize over the distribution associated with unobserved values, as detailed by Ilin and Raiko (2010).
PPCA corresponds to factorizing a covariance matrix. Another way to reduce the number of parameters in a continuous Gaussian model is to use sparse inverse covariance models, which, when combined with mixture models and EM, yield another form of clustering with correlated attributes. Edwards (2012) provides a nice introduction to graphical modeling, including mixed models with discrete and continuous components. Unlike any other treatments, he also examines graphical Gaussian models and delves further into the correspondence between a graphical model and the sparsity structure of an inverse covariance matrix. These concepts are better grasped using the “canonical” parameterization of the Gaussian distribution in terms of ![]() , and
, and ![]() , rather than the usual “moment” parameterization that uses the mean
, rather than the usual “moment” parameterization that uses the mean ![]() and covariance matrix
and covariance matrix ![]() .
.
LSA was introduced by Deerwester, Dumais, Landauer, Furnas, and Harshman (1990). pLSA has its origins in Hofmann (1999). Latent Dirichlet allocation (LDAb) was proposed in Blei et al (2003). The highly effective “collapsed Gibbs sampling” approach for LDAb was proposed by Teh et al. (2006), who also extended the concept to variational methods. Rather than applying LDAb naïvely when looking for trends over time, Blei and Lafferty (2006)’s dynamic topic models treat the temporal evolution of topics explicitly; they examined topical trends in the journal Science. Griffiths and Steyvers (2004) used Bayesian model selection to determine the number of topics in their LDAb analysis of Proceedings of the National Academy of Science abstracts. Griffiths and Steyvers (2004) and Teh et al. (2006) give more details on the collapsed Gibbs sampling and variational approaches to LDAb. Hierarchical Dirichlet processes (Teh et al., 2006) and related techniques offer alternatives to the problem of determining the number of topics or clusters in hierarchical Bayesian models. These technically sophisticated methods are quite popular, and high-quality implementations are available online.
Logistic regression is sometimes referred to as the workhorse of applied statistics; Hosmer and Lemeshow (2004) is a useful resource. Nelder and Wedderburn (1972)’s work led to the generalized linear modeling framework. McCullagh (1980) developed proportional odds models for ordinal regression, which are sometimes called ordered logit models because they use the generalized logit function. Frank and Hall (2001) showed how to adapt arbitrary machine learning techniques to ordered predictions. McCullagh and Nelder (1989)’s widely cited monograph is another good source for additional details on the framework of generalized linear models.
Tibshirani (1996) developed the famous “Least Absolute Shrinkage and Selection Operator,” also known as the LASSO; while Zou and Hastie (2005) developed the “elastic net” regularization approach.
Kernel logistic regression transforms a linear classifier into a nonlinear one, and probabilistic sparse kernel techniques are attractive alternatives to support vector machines. Tipping (2001) proposed a “relevance vector machine” that manipulates priors on parameters in a way that encourages kernel weights to become zero during learning. Lawrence, Seeger, and Herbrich (2003) proposed an “informative vector machine,” which tackles the problem with a fast, sparse Gaussian process method in the sense of Williams and Rasmussen (2006). Zhu and Hastie (2005) formulated sparse kernel logistic regression as an “import vector machine” that uses greedy search methods. However, none of these approach the popularity of Cortes and Vapnik (1995)’s support vector machines, perhaps because their objective functions are not convex, in contrast to that of an SVM (and L2 regularized kernel logistic regression). Convex optimization problems have a single minimum in the loss function (maximum in the likelihood). When probabilities are needed from an SVM, Platt (1999) shows how to fit a logistic regression to the classification scores.
Some techniques for smoothing n-grams arise from applying prior distributions to the parameters of the model’s conditional probabilities. Others come from different perspectives—such as interpolation techniques, where weighted combinations of lower order n-grams are used. Good-Turing discounting (Good, 1953) (coinvented by Alan Turing, one of the fathers of computing), and Witten–Bell smoothing (Witten & Bell, 1991) are based on these ideas. Brants and Franz (2006) discuss the massive Google n-gram collections mentioned in Section 9.8; they are available as a 24 GB compressed text file from the Linguistic Data Consortium.
Rabiner and Juang (1986) and Rabiner (1989) give a classic introduction and tutorial respectively on Hidden Markov Models. They have been used extensively for decades in speech recognition systems, and are widely applicable to many other problems. The human genome sequencing project stretched from around 1990 to the early 2000s (International Human Genome Sequencing Consortium, 2001; Venter et al., 2001), and spawned a surge of activity in recognizing and modeling genes in genomes using hidden Markov models (Burge & Karlin, 1997; Kulp, Haussler, Rees, & Eeckman, 1996)—a particularly impressive and important application of data mining. Murphy (2002) is an excellent source for details on how dynamic Bayesian networks extend hidden Markov models.
Lafferty, McCallum, and Pereira (2001) is a seminal paper on conditional random fields; Sutton and McCallum (2006) is an excellent source of further details. Sha and Pereira (2003) present the global feature vector view of conditional random fields. Our presentation synthesizes these perspectives. The original application was to sequence labeling problems, but they have since become widely used for many sequence processing tasks in data mining. Kristjansson, Culotta, Viola, and McCallum (2004) examined the specific problem of extracting information from email text. The Stanford Named Entity Recognizer is based on conditional random fields; Finkel, Grenager, and Manning (2005) give details of the implementation.
Markov logic networks (Richardson & Domingos, 2006) provide a way to create dynamically instantiated Markov random fields from programs encoded using weighted clauses in first-order logic. This approach has been used for collective or structured classification, link or relationship prediction, entity and identity disambiguation, among many others, as described in Domingos and Lowd (2009)’s textbook.
Software Packages and Implementations
Implementations of principal component analysis, Gaussian mixture models, and hidden Markov models are widely available in many software packages. MatLab’s statistics toolbox, e.g., has implementations of principal component analysis and its probabilistic variant based on the methods we have discussed, and also contains implementations of Gaussian mixture models and all the canonical hidden Markov model manipulations.
Kevin Murphy’s MatLab-based Probabilistic Modeling Toolkit is a large, open source collection of MatLab functions and tools. The software contains implementations for many of the methods we have discussed here, including code for Bayesian network manipulation and inference methods.
The Hugin software package from Hugin Expert A/S and the Netica software from Norsys are well-known commercial software implementations for manipulating Bayesian networks. They contain excellent graphical user interfaces for interacting with these networks.
The BUGS (Bayesian inference Using Gibbs Sampling) project has created a variety of software packages for the Bayesian analysis of complex statistical models using Markov chain Monte Carlo methods. WinBUGS (Lunn et al., 2000) is a stable version of the software, but the more recent OpenBUGS project is an open source version of the core BUGS implementation (Lunn, Spiegelhalter, Thomas, & Best, 2009).
The VIBES software package (Bishop, Spiegelhalter, & Winn, 2002) allows inference in graphical models using variational methods. Microsoft Research has created a programming language known as infer.net that allows one to define graphical models and perform inference in them using variational methods, Gibbs sampling or another message passing method known as expectation propagation (Minka, 2001). Expectation propagation generalizes belief propagation to distributions and models beyond the typical discrete and binary models used to create Bayesian networks. John Winn and Tom Minka at Microsoft Research have been leading the infer.net project.
The R programming language and software environment was created for statistical computing and visualization (Ihaka & Gentleman, 1996). It has its origins at the University of Auckland, and provides an open source implementation of the S programming language from Bell Labs. It is comparable to well known commercial packages such as SAS, SPSS, and Stata, and contains implementations of many classical statistical methods such as generalized linear models and other regression techniques. Since it is a general purpose programming language there are many extensions and implementations of the models discussed in this chapter available online. Historically Brian D. Ripley has overseen the development of R. Ripley is now retired from Oxford University where he was a statistic Professor. He is the coauthor of a number of books on S programming (Venables & Ripley, 2000, 2002) and an older but very high-quality textbook on pattern recognition and neural networks (Ripley, 1996).
The MALLET Machine Learning for Language Toolkit (McCallum, 2002) provides excellent Java implementations of latent Dirichlet allocation and conditional random fields. It also provides many other statistical natural language-processing methods ranging from document classification and clustering to topic modeling, information extraction, and other machine learning techniques frequently used for text processing.
The open source Alchemy software package is widely used for Markov logic networks (Richardson & Domingos, 2006).
Scikit-learn (Pedregosa et al., 2011) is a rapidly growing Python-based set of implementations of many machine learning methods. It contains implementations of many probabilistic and statistical methods for classification, regression, clustering, dimensionality reduction (including factor analysis and PPCA), model selection and preprocessing.
9.10 Weka Implementations
BayesNet (Bayesian networks without hidden variables for classification)
A1DE and A2DE (in the AnDE package)
• Conditional probability models
LatentSemanticAnalysis (in the latentSemanticAnalysis package)
ElasticNet (in the elasticNet package)
KernelLogisticRegression (in the kernelLogisticRegression package)
EM (clustering and density estimation using the EM algorithm)

