15
Application of Deep Learning Algorithms in Medical Image Processing: A Survey
Santhi B.*, Swetha A.M. and Ashutosh A.M.
SASTRA Deemed University, Thanjavur, Tamil Nadu, India
Abstract
Deep learning (DL) in medical image processing (MIP) and segmentation for a long time and it continues to be the most popular and not to mention a powerful technique, given its exceptional capability in image processing and classification. From fledgeling DL models like convolutional neural networks (CNNs), which is, by far, the best rudimentary yet convoluted model for image classification, to complex algorithms like transfer learning which involves model construction on top of state-of-the-art pre-trained classifiers, DL has established itself as a capable and potential technique for medical imaging processing. Prior to the development of DL models, MIP or image processing, in general, was restricted to edge-detection filters and other automated techniques. But the advent of artificial intelligence (AI) and, along with it, the instances of ML and DL algorithms changed the facet of medical imaging. With adequate dataset and proper training, DL models can be made to perfect the task of analyzing medical images and detecting tissue damage or any equivalent tissue-related abnormality with higher precision.
This paper summarizes the evolution and contribution of various DL models in MIP over the past 5 years. This study extracts the fundamental information to draw the attention of researchers, doctors, and patients to the advancements in AI by consolidating 80 papers that categorize five areas of interest. This comprehensive study increases the usage of AI techniques, highlights the open issues in MIP, and also creates awareness about the state of the art of the emerging field.
Keywords: MIP, deep learning, classification, segmentation, feature engineering
15.1 Introduction
In the emerging times, artificial intelligence (AI) has extended into all sectors of knowledge like government, security, medical, and agriculture through automations in different domains. AI was founded by a group of researchers in the early 1950s and has since gained popularity (after having faced inexorable winters), with the increase in technology. Machine learning (ML) and deep neural networks are the two branches of AI that have revolutionized AI.
Medical imaging refers to the myriad of procedures involved in procuring digital images of a subject’s internal organs for the diagnosis of associated ailments and to also identify and detect changes in the functioning of inter tissues. With the advancements in the field of digital imaging, detailed multi-planar vision of the interiors of the human body with exceptional resolution has been made possible. Rapid developments in the domain of image processing including the development of techniques such as image recognition have enhanced the approach of studying and analyzing different body parts and diseases. The thriving domain of digital image processing has amplified the accuracy of diagnosing a disease and also in detecting the expanse of damage to tissues as a result of the ailment. Medical imaging uses non-invasive approaches to obtain segmented images of the body, which are transmitted to computers as signal and, are converted to digital images on reception for the detection of anomalies and defects.
Several techniques exist for medical imaging which include both organic as well as radiological imaging techniques together with thermal, magnetic, isotopic, and sonogram-based imaging among a few. Though the usage of ML in industries is increasing, deep learning (DL) being the subset of ML, outshines the traditional rule-based algorithms in extracting the features and domain expertise. In traditional algorithms, the features are identified by the experts to minimize the complexity of the intelligence. The DL algorithms achieve higher precision and accuracy by learning incrementally to pick out the important features in the enormous data for accurate predictivity analysis.
A parallel and profound development in the domain of DL has changed the way in which medical images are processed and analyzed. With the overwhelming inflow of DL algorithms, computer models are being trained to study and understand the subtleties of the human body. These DL models are trained on a set of available medical images [38] that gives the model a basic overview of the internal framework of the human body. Increasing advancements in DL algorithms have given birth to models that detect tissue damage and other internal ailments with exceptional accuracy and precision. These developments have reshaped the field of medicine and are crucial for better and early diagnosis.
15.2 Overview of Deep Learning Algorithms
The ML algorithms in medical image processing (MIP) can be classified as supervised and unsupervised deep neural networks.
15.2.1 Supervised Deep Neural Networks
A neural network is called supervised if the expected output is predefined and during the training phase the neurons in the networks are trained accurately with the target output. Regularization and back propagation can be used on the network to train the model so that the output is almost closer to the expected output with minimum error or loss. The most frequently used supervised deep neural networks in MIP are convolutional neural network (CNN), transfer neural network, and recurrent neural networks (RNNs).
15.2.1.1 Convolutional Neural Network
CNN [4] is the most commonly used composite deep feedforward neural network used in image classification and segmentation. CNN can extract higher level features from the image accurately which signifies that this model is the core component in image classification problems. CNN architecture consists of convolution layer, pooling layer, and fully connected layer.
Convolutional layer: This layer applies the convolution on the input image with filters to extract the feature map from the images, constructing an output feature map of the same size as the filter. The filter is moved over the image based on the stride assigned and the feature map of the entire image is calculated using convolution function.
The ReLU activation function is applied on the hidden layer neurons to impose nonlinearity in the model. The ReLU function is given by, f(x) = max(0,x), for any positive value of x.
Pooling layer: This layer downsamples the feature map into lower dimension by preserving the important features in the input feature. The pooling can be divided as max pooling and average pooling, the former takes the high value in the window as the output feature and the later calculates the average of all the values in the window to be the output feature value.
Fully connected layer: The fully connected layer performs the classification based on the features extracted in the previous layers. The softmax activation function is applied on the output layer so that the output is classified in the probability space.
15.2.1.2 Transfer Learning
Models like CNN require a lot of data for training the model to get better accuracy, but in real-time medical applications, the publicly available data is very minimal which restricts the performance of a model that is built from scratch. In case of less training samples, models like transfer learning [75] can be trained since these models are already trained with the huge collection of data, reducing the training time of the model. The weights of the pre-trained models in transfer learning are loaded except the weights in the output layer. The model is trained by analyzing the patterns in the training data and the new custom layer can be added at the end of the model which best fits the classification or feature extraction problem. Some of the most high performance transfer learning networks are VGGNet, ResNet, AlexNet, DenseNet [65], Inception, and GoogleNet. These networks are the collection of series of CNN feedforward networks with a variable number of layers and parameters.
AlexNet consists of five convolutional layers while VGGNet and GoogleNet have 19 and 22 layers, respectively. However, transfer learning is simply not about stacking the layers to increase the network. As the depth of the network increases, there is a higher chance of degrading the performance of the model due to vanishing gradient problems. ResNet [19] circumvents the problem of vanishing gradients by skipping the intermediate connections and performing identity feature mapping with their outputs and the stacked layers.
15.2.1.3 Recurrent Neural Network
RNN [68] is a deep neural network that takes into consideration the historical information, i.e., in other words, it uses previous outputs as inputs while using the hidden states to produce a series of output vectors. RNN finds its purpose in image recognition, speech recognition, etc. Unlike a vanilla neural network, RNN remembers prior inputs that form its internal memory and is thereby best-suited for modeling sequential data. It copies the output from the previous information cycle and pushes it into the network along with the current input after assigning them weights. The weights are assigned through backpropagation and gradient descent. The disadvantage of RNNs is that it has a short-term memory which prevents proper tuning of weights via backpropagation. However, this disadvantage can be overcome using LSTM.
15.2.2 Unsupervised Learning
The neural network that performs some task without any pretext labels is called unsupervised learning. The images that are similar are grouped together into the same class. Some of the most frequently used unsupervised deep neural networks are autoencoders and generative adversarial networks (GANs).
15.2.2.1 Autoencoders
Autoencoder is an unsupervised neural network model that effectively compresses data and encodes it to fit the imposed network bottleneck, thereby providing a compressed representation of the input information. This encoded data can be decoded using the same technique to obtain a data representation that is as accurate as the input. An encoder has four stages namely encoder, bottleneck, decoder, and reconstruction error. The compressed data is its lowest possible dimensional representation. The reconstruction error is the difference between the decoded representation of the compressed data and the original one. This error factor helps the model to better adapt to the original data via backpropagation, thereby reducing the error margin. Autoencoders are used in image denoising, anomaly detection, noise reduction, etc.
15.2.2.2 GANs
GANs [31] are a class of powerful neural network models. They are used to generate example data when the dataset is too little to be used for model training, thereby increasing classifier accuracy. They are also used for increasing image resolutions and morphing audio from one speaker to another. GANs function with two sub-models called generators and discriminators. A generator is a model that generates examples that are drawn from the original data. A discriminator labels the generated examples as real or fake. Both the models are trained simultaneously and the generator is constantly updated on how well it performed. When a generator model fails to fool the discriminator, the model parameters of the generator are tuned to enhance its performance.
15.3 Overview of Medical Images
The medical images are the tasks that are used to create the images of different parts of the anatomy for the sake of treatment and diagnostics of the patient’s health. Medical images help practitioners to view the biological changes in the patient’s body and diagnose the condition with more accurate and suitable treatment decisions. Figure 15.1 shows various radiographic imaging techniques used in medical imaging.
15.3.1 MRI Scans
MRI (magnetic resonance imaging) [17] is a procedure used to visualize a detailed cross-section of the patient’s body. The imaging technique uses powerful magnetic fields coupled with radio waves to generate a circumstantial image of the patient’s internals, thereby making it easy to detect and locate abnormalities. The fluctuating magnetic field stimulates the protons found in the living tissue and aligns them along the field direction. The radio waves interact with these aligned protons and produce resonating signals which are tapped by the receiver to be displayed as computer-processed images of the body tissue. The MRI scan images show the desired body part with greater clarity accentuating the affected region. This makes it easy for physicians to locate any irregularity, which would have otherwise been difficult to detect.
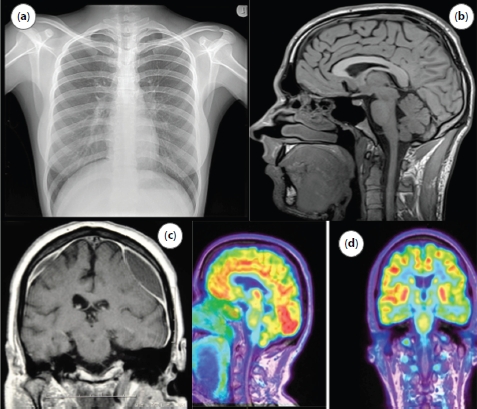
Figure 15.1 Digital medical images: (a) X-ray of chest, (b) MRI imaging of brain, (c) CT scan of brain, and (d) PET scan of brain.
15.3.2 CT Scans
CT (computed tomography) [13] scans use a rotating X-ray generator to generate a high-quality fragmented image of the body. This method is used to diagnose disorders associated with bone-marrow, blood vessels, and other soft tissues. The machine captures X-ray images of the body from different angles, which are later combined to produce a three-dimensional image of the body. CT scans also enable multiplanar views of the body of late, which helps us view reformatted images in traverse as well as the longitudinal plane. A patient is intravenously administered with contrast agents which enhances the contrast of body fluids. These agents are used to better visualize blood vessels. The threat associated with this technique is that it uses ionizing radiation which may prove to be carcinogenic.
15.3.3 X-Ray Scans
X-ray scans are critical for diagnosing bone-related injuries like fractures, dislocation, or misalignment, and tumors. An x-ray generator shoots high-frequency x-ray radiations which are received by the detector on the opposite end as it passes through the patient’s body. Bones have high calcium density hence absorb most of the x-ray radiations while soft tissues allow the rays to pass through easily. This helps form a well-structured definition of the bone framework. To prevent undesirable radiation exposure some body parts can be covered with lead aprons. Apart from bone injuries x-rays are used to detect pneumonia and are used in mammograms to detect breast tumors.
15.3.4 PET Scans
PET (positron emission tomography) scans are used to check the viability and the functionality of living tissue. The patient is administered with a radioactive drug called tracer either orally or via injection. Organs that have abnormalities usually display high chemical activity. The tracer cumulates in these organs insinuating associated irregularities which help the physicians detect ailments even before it shows up on other scans.
15.4 Scheme of Medical Image Processing
The medical images of the human body that are collected from various radiographic imaging techniques are required to be handled and processed by computers so that the DL models can classify the images as normal or abnormal without any human intervention. The medical images processing encompasses five major areas [12]: image formation, enhancement, analysis, visualization, and management, as shown in Figure 15.2.
15.4.1 Formation of Image
To analyze the images quantitatively, a good understanding about the projection and digitalization of the images is very essential. There are different modalities of images for MIP like CT, PET, MRI, X-rays, and ultrasound, each of which has different qualities. The image formation involves acquisition of raw images and then digitalizing the images into matrix for further pre-processing and analysis.
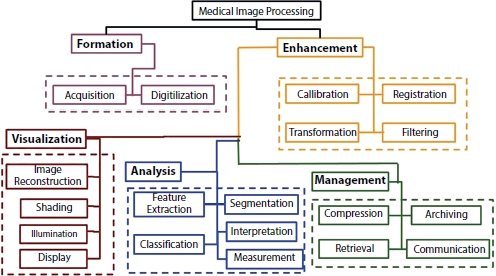
Figure 15.2 Scheme of image processing [12].
15.4.2 Image Enhancement
The main idea behind enhancement is to unwrap the hidden features in the images that are more suitable for classification. Image enhancement involves transforming the images into spatial or frequency domain in order to improve the interpretability of these images. The technique in which the pixels from an image are utilized to calibrate it is called spatial domain technique. For sharpening and smoothening the images, frequency domain technique is used. The images are aligned to analyze the temporal changes in multiple images in the image registration technique. These techniques allow elimination of noise and heterogeneity in the images by improving the most relevant features that are required for the analysis.
15.4.3 Image Analysis
Preliminary to image classification, the feature extraction and segmentation are the most important steps in analyzing the nature of the image for classification. Segmentation [3] is the partitioning of different anatomical structures into contours. The accuracy of the classification relies on accuracy of the segmentation. Feature extraction deals with extracting relevant attributes for image recognition and interpretation. The image classification is the procedure of designating labels to the object based on the information extracted from the image.
15.4.4 Image Visualization
This process renders the images from initial or intermediate steps for visualizing the anatomical images.
15.5 Anatomy-Wise Medical Image Processing With Deep Learning
MIP is important in diagnosis of the patient’s health condition by the practitioners. The medical imaging can be of multiple modalities like CT, MRI, mammograms, X-rays, and ultrasound. Table 15.1 gives the summary of the datasets used in the anatomy-wise analysis of different papers. Analyzing these digital images can be beneficial in segregating the normal patients from the abnormal patients who need the attention in diagnosis of the medical condition. Some of the medical diagnoses that can be made by studying the scans of the patients’ anatomical structure are detection and segmentation of brain tumor, breast cancer, lung nodules, pulmonary artery, diabetic retinopathy, etc. Figure 15.3 shows the collection of papers that are published in different anatomical-wise medical image classification using DL, and it is evident that the evolution of DL in medical image has rapidly increased from 2016 to 2020. Figure 15.4 compares the number of publications on different analysis from the digital medical images.
Table 15.1 Summary of datasets used in the survey.
| S. no. | Dataset | Dataset abbreviation | Dataset description |
| Brain tumor | |||
| 1. | BRATS 2015 | Multimodal Brain Tumor Image Segmentation (BRATS) | The Dataset is prepared from NIH Cancer Imaging Archive (TCIA) and contains 300 high- and low-grade glioma cases. |
| 2. | T1 -W CEMRI | T1-weighted dynamic contrast-enhanced MRI (CEMRI) | The T1 weighted CEMRI dataset is obtained from the NBDC database archive and contains structural images involving clinical information on patients with unipolar and bipolar diseases and schizophrenia. |
| 3. | FLAIR | Fluid-attenuated inversion recovery (FLAIR) | This dataset contains structural MRI images for medical image visualization and processing collected as part of a neurohacking course available in coursera consisting of 176 cases. |
| Lung nodule cancer dataset | |||
| 1. | LIDC/IDRI | Lung Image Database Consortium image collection (LIDC-IDRI) | The dataset comprises of CT scans with annotated lesions of Lung cancer thoracic regions. |
| 2. | 18F-FDG PET/CT | PET with 2-deoxy-2-[fluorine-18]fluoro-D-glucose integrated with computed tomography (18F-FDG PET/CT) | The dataset is the combined procurement of PET and CT scans of the thoracic region. |
| Breast cancer | |||
| 1. | DDSM | Digital Database for Screening Mammography (DDSM) | The DDSM dataset comprises 2,620 samples of mammogram images of the patients annotated as normal, benign and malignant. |
| 2. | WBCD | Wisconsin Breast Cancer Dataset | The dataset consists of 701 instances of patients with solid breast mass. |
| 3. | MIAS | mini-Mammographic Image Analysis Society | MIAS dataset consists of 322 mammogram images from 161 patients. |
| Heart disease prediction | |||
| 1. | PTB Diagnostic ECG Database | Physikalisch Technische Bundesanstalt (PTB) Diagnostic ECG Database | The dataset consists of 549 instances ECG signals from 290 patients with normal and abnormal behavior in the heart under different age groups. |
| 2. | PhysioNet Public Database | BIDMC Congestive Heart Failure Database | The dataset consists of ECG recordings of severe congestive heart failure from 15 patients. |
| COVID-19 prediction | |||
| 1. | COVID-CT-Dataset | Chest CT scans | The dataset consists of 749 images of patients with COVID and nonCOVID cases. |
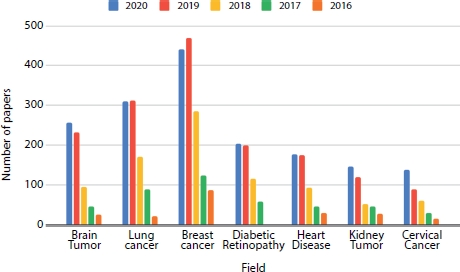
Figure 15.3 Anatomy-wise breakdown of papers in each year (2016–2020).

Figure 15.4 Year-wise breakdown of papers (2016–2020) based on the task.
15.5.1 Brain Tumor
The brain tumor is the erratic widening of cells in the brain, which can be infectious or benign. The cells that are homogeneous in composition and are not active are called benign tumor cells and the ones that are heterogeneous in composition with the presence of active cells that grow uncontrollably in the brain are categories as malignant. MRI scans can detect such tumor conditions in the brain and many more abnormal mass growths like cysts and aneurysms and swelling.
Authors in [76] proposed multi-level Gabor wavelet filters for abstracting the relevant features from the MR images and then SVM is trained on the extracted features. Conditional random fields are applied on the output of the SVM to segment the tumor. Authors in [33] proposed a skull stripping technique, fuzzy hopfield neural network for segmenting and detection of tumor in the brain images. These works use traditional feature extraction methods like wavelet filters, watershed algorithms [2], thresholding, and active contour model. To accurately segment the features, recent works focus on DL networks like CNN or transfer learning models [1] for feature extraction from the images.
Table 15.2 Summary of papers in brain tumor classification using DL.
| S. no. | Author | Data | Model | Description |
| 1. | [76] | BRATS 2015 | CRF, SVM | Multi-level Gabor wavelet filters for segmenting the important features from the MR images and then SVM is trained on the extracted features. |
| 2. | [33] | BRATS 2015 | Fuzzy Hopfield neural network | Fuzzy hopfield neural network for segmenting the tumors. |
| 3. | [14] | BRATS 2015 | U-Net | U-Net-based CNN for segmenting the tumor cells from brain images. |
| 4. | [49] | BRATS 2015 | Watershed Algorithm for Segmentation | The MRI image is transformed into the grayscale digital image (GDI) and the segmentation technique like watershed algorithm is applied on the GDI image to extract the portion of the image containing the tumor. |
| 5. | [1] | BRATS 2015 | Phase-1: CNN and SVM Phase-2: VGG, AlexNet | The phase-1 classification which includes feature extraction and classification of brain images as ordinary and abnormal is carried out using CNN and SVM algorithm. The phase-2 segmentation involves localization of the tumor in the abnormal brain images using three CNN-transfer learning techniques. |
| 6. | [6] | BRATS 2015 | RF | Classification of brain images as tumorous or not using voting average-based random forest classifier. |
| 7. | [57] | MRI | Probabilistic Neural Network | Gray-Level Co-occurrence Matrix (GLCM) is used to abstract the features from the MRI images which increases the accuracy and reduces the time for prediction. The PNN classifier is used to find whether the image contains glioblastoma or not. |
| 8. | [27] | BRATS 2015 | GMM, Fuzzy Means Clustering | Wavelet transform removes noises in the BMRI images and then active contours and GMM are applied on the image to perform skull stripping on the brain images. |
| 9. | [71] | SAR | PNN-RBF | The brain images are classified using PNN-RBF classifier and the tumor is segmented and clustered using K-means clustering. |
| 10. | [53] | T1-W CEMRI | Hybrid Kernelbased Fuzzy C-Means clustering -Convolutional Neural Network |
|
| 11. | [69] | BRATS 2015 | SVM, fuzzy clustering and probabilistic local ternary patterns |
|
| 12. | [51] | BRATS 2015 | CNN, WT | The brain image features are extracted using wavelet transform and then CNN is applied on the extracted features to classify the brain as tumorous or not. |
| 13. | [27] | T1CE, FLAIR | DL model | Detection of meningiomas using multiparametric deep learning model (DLM). |
Table 15.2 presents the state of the art of various DL methods that are used in the classification of Brain tumors using MRI scans.
15.5.2 Lung Nodule Cancer Detection
Lung nodules are small masses in lung tissues which can either be cancerous or not. CT scans are an effective way of detecting the lung nodules at early stages. The CAD-based technique can be categorized as the detecting system, which segments the nodules from the tissue, and the diagnostic system which classifies the segmented nodule as benign or malignant. Identifying such nodules through CT scans manually requires experience in the field, and it may also lead to negligence and can cause severe problems.
Table 15.3 Paper summary—cancer detection in lung nodule by DL.
| S. no. | Author | Data | Model | Description |
| 1. | [62] | LIDC/IDRI | CNN | CNN-based cancer malignancy classification. |
| 2. | [67] | LIDC/IDRI | CNN, DNN, and SAE |
|
| 3. | [39] | LIDC/IDRI | ResNet | Transfer learning-based classification model to classify the CT scans to predict if lung image contains cancer or not. |
| 4. | [74] | 18F-FDG PET/CT | CNN and ML models |
|
| 5. | [21] | LIDC/IDRI | CNN | The images obtained by the median intensity projection are concatenated and data augmentation is fitted on the images to increase the sample size, and the features from the images are extracted using CNN model and then Gaussian process model is applied on the extracted features to predict the malignancy score. |
| 6. | [66] | LIDC/IDRI | U-Net | U-Net architecture for classifying the nodule is cancerous or not. |
| 7. | [22] | CT, MRI | GAN | Semi-supervised model, GAN is proposed to extract the nodule information from CT and MRI scans of the patients to detect if the nodule in the lung is benign or malignant. |
| 8. | [80] | LIDC/IDRI | AgileNet | Hybrid of LeNet and AlexNet is proposed to classify the nodules. |
| 9. | [26] | LIDC/IDRI | Optimal Deep Neural Network (ODNN) and Linear Discriminant Analysis (LDA) | The computation time and cost during classification can be reduced using LDA as the feature selection technique, the CT lung images are classified as normal, benign, and malignant based on the features that are extracted from the ODNN approach an optimized using Modified Gravitational Search Algorithm (MGSA) for detect the lung cancer classification. |
| 10. | [28] | LUNA16, DSB2017 | 3D-Deep Neural Network | Model consists of two modules. The former is a 3D region proposal network for nodule detection. The latter selects the most relevant five nodules based on the detection confidence. Cancer probabilities are evaluated and combine them with a leaky noisy or gate. |
| 11. | [20] | Lung Nodule Analysis 2016, Alibaba Tianchi Lung Cancer Detection | U-Net | The presence of nodules in the lung images does not conclude that the lung is cancerous, so a 3D DL model, U-Net, is proposed to detect the nodules in the lung CT image and then predicting if the scan is abnormal by classifying the nodules into the five class probability spaces. |
| 12. | [79] | LIDC/IDRI | Multi-view knowledgebased collaborative | Multi-view knowledgebased collaborative model is proposed using ResNet-50 to separate the malignant nodules from benign |
| 13. | [61] | LIDC/IDRI | Deep hierarchical semantic convolutional neural network | Deep hierarchical semantic CNN-based model to forecast the less important features and relevant features to detect if the nodule is malignant. |
| 14. | [8] | Chest X-Ray, LIDC-IDRI | Modified AlexNet (MAN) |
|
| 15. | [67] | LIDC/IDRI | CNN, DNN and SAE |
|
DL is the effective way which increases the precision and accuracy of classification and detection of the infectious nodules from the CT scans of the patients. It is perceptible that many DL methods like CNN [62], CNN-based transfer learning [39], and unsupervised DL GANs [22] extract the features from the CT images more accurately than any other means.
Table 15.3 presents the state of the art of various DL methods that are used in the detection of Lung nodule cancer using CT/MRI scans.
15.5.3 Breast Cancer Segmentation and Detection
The second major reason of death among women is breast cancer and nearly 8% of the women population gets afflicted with it. Early detection and treatment of breast cancer can elevate the process of treatment and save the life of many patients. DL models are proven to be one the effective method for detecting the abnormal tissues in the mammograms images.
Table 15.4 Paper summary—classification of breast cancer by DL.
| S. no. | Author | Data | Model | Description |
| 1. | [40] | DDSM | Maximum difference feature selection (feature extraction technique) | The proposed maximum difference feature selection involves extracting the relevant tissue information from the mammograms for early detection of breast cancer. |
| 2. | [41] | DDSM | Neural network | CAD-based approach to classify the breast mammogram images as cancer and normal by extracting the tissue features and applying neural network on the extracted features. |
| 3. | [42] | DDSM | ANN | Intensity-based feature extraction technique to segment the features from the digital mammograms and classifying the images as normal and abnormal using ANN. |
| 4. | [2] | WBCD | Deep-belief network | Construction of backpropagation neural network and deep belief network path to initialize the weights in the network |
| 5. | [44] | MIAS | ANN, LDA, Naive Bayes | Various features like Wavelet Packet Transform (WPT), Local Binary Pattern (LBP), Entropy, Gray Level Co-Occurrence Matrix (GLCM), Discrete Wavelet Transform (DWT), Gray Level Run Length Matrix (GLRLM), Gabor transform, Gray Level Difference Matrix (GLDM), and trace transform are extracted from histogram and the important features are selected using ANOVA and are fed to the classifiers for classifying the breast density as two-class or three-class. |
| 6. | [43] | MIAS | Graph-cut segmentation technique | Involves segmenting the breast regions from the image and finding the percentage of fat and dense tissue in the breast density. |
| 7. | [56] | MIAS | RNN, SVM | Involves RNN for selecting the features and SVM to classify the image as normal or abnormal. |
| 8. | [25] | MIAS | GoogleNet, Visual Geometry Group Network (VGGNet) and Residual Networks (ResNet) | Pre-trained CNN architecture for classification of mammogram images into normal or abnormal. |
| 9. | [60] | CBIS- DDSM | Resnet50 and VGG16 | Pre-trained CNN architecture for classification of mammogram images into normal or abnormal. |
Table 15.4 presents the state of the art of various DL methods that are used in the classification of breast cancer using mammography.
15.5.4 Heart Disease Prediction
Heart disease in a nutshell is a condition that affects the heart, which includes diseases like blood vessel diseases, such as coronary artery disease, arrhythmias (heart rhythm problems), and congenital heart defects (heart defects by birth). According to the World Health Organization (WHO) heart diseases are the leading causes of deaths among the population, taking the lives of 17.9 million people world-wide each year. Therefore, an early prediction of potential heart disease is the most vital research in the medical analysis which can eventually help doctors get insight into heart failures and change their treatment accordingly. However, it is difficult to manually find the risk factors of heart disease. Modern day DL techniques [16] have proven results in determining the knowledge about the factors leading to disease and medical data exploration in predicting the disease as early as possible to avoid any critical health condition.
Table 15.5 Paper summary on heart disease prediction using DL.
| S. no. | Author | Data | Model | Description |
| 1 | [15] | Health system’s EHR | RNN | The relation between the time-stamped events with the observation window of 12 to 18 months were detected using RNN models with Gated Recurrent Units (GRUs). |
| 2 | [52] | Physikalisch-Technische Bundesanstalt diagnostic ECG database | CNN |
|
| 3 | [77] | PhysioNet public database | MS-CNN |
|
| 4 | [24, 34] | Cleveland heart disease dataset | DNN multilayer perceptron | Deep neural model is proposed for classification of the heart disease. |
| 5 | [47] | PhysioNet public database | CNN | A 16-layered CNN model is adapted to classify cardiac arrhythmia cases from the long-duration ECG signals. |
| 6 | [48] | PhysioNet public database | CAE-LSTM |
|
| 7 | [29, 30] | Cleveland heart disease dataset | ANN, DNN |
|
| 8 | [18] | EHR (electronic health records) | LSTM | LSTM-based model for early treatment of heart failure is a superior and novel approach compared to other methods in HF diagnosis. |
| 9 | [45] | TB diagnostic ECG and Fantasia Databases, Petersburg Institute of Cardiological Technics 12-lead Arrhythmia Database, PTB Diagnostic ECG Database and BIDMC Congestive Heart Failure Database | CNN, LSTM | ECG signals are classified into CAD, MI, and CHF conditions using CNN, followed by combined CNN and 16-layered LSTM models. |
| 10 | [9] | Health Insurance Review and Assessment Service (HIRA) - National Patients Sample (NPS) | RNN-based RetainEX | RetainVis is an analytical tool built for interpretable and interactive RNN-based models called RetainEX and visualizations for users’ exploration of EMR data in the context of prediction tasks. |
| 11 | [11, 64] | CPC of Shanxi Academy of Medical Sciences | CNN-RNN | The features of the ECG signals are extracted using the CNN and the feature sets are formed by combining the clinical features which are fed to an RNN for classification. |
| 12 | [10] | External echocardiogram videos set from Cedars-Sinai Medical Center, internal set from Stanford university | EchoNet- Dynamic | The model surpasses the performance of human experts in the critical tasks of segmenting the left ventricle, estimating ejection fraction and assessing cardiomyopathy. |
Table 15.5 presents the state of the art of various DL methods that are used in the prediction of heart disease using ECG/EEG signals.
15.5.5 COVID-19 Prediction
COVID-19 (coronavirus disease) which originated as a mere endemic in the capital city of Wuhan, Hubei Province, China, has spread across the globe over the course of a year affecting millions of people, majority of whom have succumbed to it. The current diagnosis methods for the detection of COVID-19 patients include laboratory examinations, nasopharyngeal, and oropharyngeal swab tests. Swab tests are done by inserting a long swab into the person’s nostril to collect sample secretions and cells from the nasal passage that connects it to the hind of the throat. The secretions are later tested for traces of COVID-19 genetic strains using a PCR (polymerase chain reaction) assay.
Despite the development of standard quantitative RT-PCR tests, they have been regarded as unreliable for COVID-19 diagnosis as they showed an accuracy of only 56%. CT scans have shown higher detection accuracy (around 88%) as the imaging findings of COVID-19 resembles that of MERS-CoV and SARS-CoV. The CT scans of the thoracic cavity can be used to detect the severity of infection among patients. The material-enhanced CT scans show alveolar damages which are characteristic of the coronavirus family. Hence, for early diagnosis, chest CT scans have been considered as the best method.
Table 15.6 presents the state of the art of various ML and DL methods [72] that are used in the prediction of COVID-19 patients from their chest x-ray and outlines the critical role of deep neural networks in processing the chest X-ray images.
Table 15.6 COVID-19 prediction paper summary.
| S. no. | Author | Dataset | Model |
| 1. | [78] | CT Scan | ResNet |
| 2. | [70] | X-ray | Random Forest, REP Tree, Logistic, Random Tree, Simple Logistic, MLP, BayesNet, Naive Bayes |
| 3. | [23] | X-ray | DenseNet |
| 4. | [59] | X-ray | COV-ELM(ML) |
| 5. | [5] | X-ray | VGG16 |
| 6. | [37] | X-ray | SVM, KNN, RandomForest, Naïve Bayes, Decision Tree |
| 7. | [50] | X-ray | Inception V3, Xception, ResNet |
| 8. | [63] | X-ray | ResNet-18, ResNet-50, SqueezeNet, DenseNet-121 |
| 9. | [7] | X-ray | ResNet-50 |
| 10. | [73] | X-ray | Inception V3 |
| 11. | [54] | X-ray | ResNet |
| 12. | [55] | X-ray | ML Models |
| 13. | [35] | X-ray | VGG16, VGG19, MobileNet, ResNet, DenseNet-121, InceptionV3, Xception, Inception ResNet V2 |
| 14. | [36] | X-ray | ResNet-50 |
| 15. | [58] | X-ray | CNN |
| 16. | [32] | X-ray | AlexNet, VGGNet-19, ResNet, GoogleNet, SqueezeNet |
| 17. | [46] | Xray | CNN |
15.6 Conclusion
This study reviews nearly 80 papers on the topic of using DL in MIP, and it is obvious from this survey that DL has penetrated in all the aspects of the MIP. One of the biggest drawbacks in MIP is the data imbalance problem, where the training dataset is skewed over the abnormal class samples and could lead to the decrease in the performance of the model which, in turn, can lead to severe consequences if predicted falsely. Data augmentation and transfer learning mechanisms can be used as the techniques to terminate data imbalance problems. Another loophole in MIP is that not all data are publicly available. Finally, anatomy-wise models play an important part, as the algorithms need to construct the patient-specific models with minimum sized dataset and minimum user-interaction.
Although there are some challenges in MIP, from this survey, it is proved that DL models outperform medical practitioners in classification and segmentation of medical images. We envision that DL techniques can be used on various practical applications like image captioning, image reconstruction, content-based image retrievals and surgical robots using reinforcement learning providing an assistance to the medical practitioners and also helps in remote monitoring of patients.
Deep neural networks like CNN require a large and feature engineered dataset to train on and hypertuned parameters to outperform in the evaluation. There is no universal model that performs best for the whole pool of datasets available for analysis. The prediction and the performance of the model depend on the nature and quality of the dataset on which the model is trained.
References
1. Abd-Ellah, M., Awad, A., Khalaf, A., Hamed, H., Two-phase multi-model automatic brain tumour diagnosis system from magnetic resonance images using convolutional neural networks. Eurasip J. Image Video Process., 2018, 1, 2018, 10.1186/s13640-018-0332-4.
2. Abdel-Zaher, A. and Eldeib, A., Breast cancer classification using deep belief networks. Expert Syst. Appl., 46, 139–144, 2016, 10.1016/j.eswa.2015.10.015.
3. Abinaya, P., Ravichandran, K.S., Santhi, B., Watershed segmentation for vehicle classification and counting. Int. J. Eng. Technol., 5, 2, 770–775, 2013.
4. Mortazi, A. and Bagci, U., Automatically Designing CNN Architectures for Medical Image Segmentation. International Workshop on Machine Learning in Medical Imaging, 11046, pp. 98–106, 2018, https://doi.org/10.1007/978-3-030-00919-9_12.
5. Chen, A., Jaegerman, J., Matic, D., Inayatali, H., Charoenkitkarn, N., Chan, J., Detecting Covid-19 in Chest X-Rays using Transfer Learning with VGG16. CSBIO 20 Computational Systems - Biology and Bioinfromatics, pp. 93–96, 2020, https://doi.org/10.1145/3429210.3429213.
6. Anitha, R. and Siva Sundhara Raja, D., Development of computer-aided approach for brain tumor detection using random forest classifier. Int. J. Imaging Syst. Technol., 28, 1, 48–53, 2018, 10.1002/ima.22255.
7. Makris, A., Kontopoulos, I., Tserpes, K., COVID-19 detection from chest X-Ray images using Deep Learning and Convolutional Neural Networks, medRxiv 2020.05.22.20110817, 2020, https://doi.org/10.1101/2020.05.22.20110817.
8. Bhandary, A., Prabhu, G., Rajinikanth, V., Deep-learning framework to detect lung abnormality – A study with chest X-Ray and lung CT scan images. Pattern Recognit. Lett., 129, 271–278, 2020, 10.1016/j.patrec.2019.11.013.
9. Kwon, B.C., Choi, M.-J., Kim, J.T., Choi, E., Kim, Y.B., Kwon, S., Sun, J., Choo, J., RetainVis: Visual Analytics with Interpretable and Interactive Recurrent Neural Networks on Electronic Medical Records. IEEE Trans. Visual. Comput. Graphics, 25, 1, 299–309, 2018, 10.1109/TVCG.2018.2865027.
10. Ouyang, D., He, B., Ghorbani, A., Yuan, N., Ebinger, J., Langlotz, C.P., Heidenreich, P.A., Harrington, R.A., Liang, D.H., Ashley, E.A., Zou, J.Y., Video-based AI for beat-to-beat assessment of cardiac function. Nature, 580, 7802, 252–256, 2020, https://doi.org/10.1038/s41586-020-2145-8.
11. Li, D., Li, X., Zhao, J., Bai, X., Automatic staging model of heart failure based on deep learning. Biomed. Signal Process. Control, 52, 77–83, 2019, https://doi.org/10.1016/j.bspc.2019.03.009Get.
12. Deserno, T.M., Fundamentals of Biomedical Image Processing, in: Biomedical Image Processing. Biomedical Engineering, 2011, 10.1007/978-3-642-15816-2 1.
13. Mhaske, D., Rajeswari, K., Tekade, R., Deep Learning Algorithm for Classification and Prediction of Lung Cancer using CT Scan Images. ICCUBEA, 2020, 10.1109/ICCUBEA47591.2019.9128479.
14. Dong, H., Yang, G., Liu, F., Mo, Y., Guo, Y., Automatic brain tumor detection and segmentation using U-net based fully convolutional networks. Commun. Comput. Inf. Sci., 723, 506–517, 2017, 10.1007/978-3-319-60964-5_44.
15. Choi, E., Schuetz, A., Stewart, W.F., Sun, J., Using recurrent neural network models for early detection of heart failure onset. J. Am. Med. Inf. Assoc., 24, 2, 361–370, 2016, https://doi.org/10.1093/jamia/ocw112.
16. Ali, F., El-Sappagh, S., Riazul Islam, S.M., Kwak, D., Ali, A., Imran, M., Kwak, K.-S., A smart healthcare monitoring system for heart disease prediction based on ensemble deep learning and feature fusion. Inform. Fusion, 63, 208–222, 2020, https://doi.org/10.1016/j.inffus.2020.06.008.
17. Mohan, G. and Monica Subashini, M., MRI based medical image analysis: Survey on brain tumor grade classification. Biomed. Signal Process. Control, 39, 139–161, 2018, https://doi.org/10.1016/j.bspc.2017.07.007.
18. Maragatham, G. and Devi, S., LSTM Model for Prediction of Heart Failure in Big Data. J. Med. Syst., 43, 5, 111, 2019, https://doi.org/10.1007/s10916-019-1243-3.
19. He, K., Zhang, X., Ren, S., Sun, J., Deep Residual Learning for Image Recognition. Proceedings of the IEEE Computer Society Conference on Computer Vision and Pattern Recognition, 2016.
20. Huang, W. and Hu, L., Using a Noisy U-Net for Detecting Lung Nodule Candidates. IEEE Access, 7, 67905–67915, 2019, 10.1109/ACCESS.2019.2918224.
21. Hussein, S., Gillies, R., Cao, K., Song, Q., Bagci, U., TumorNet: Lung nodule characterization using multi-view Convolutional Neural Network with Gaussian Process. Proceedings - International Symposium on Biomedical Imaging, pp. 1007–1010, 2017, 10.1109/ISBI.2017.7950686.
22. Jiang, J., Hu, Y., Tyagi, N., Zhang, P., Rimner, A., Mageras, G.S., Deasy, J.O., Veeraraghavan, H., Tumor-aware, adversarial domain adaptation from CT to MRI for lung cancer segmentation. Lect. Notes Comput. Sci. (including subseries Lecture Notes in Artifical Intelligence and Lecture Notes in Bioinformatics), 11071, 777–785, 2018, 10.1007/978-3-030-00934-2_86.
23. Paul Cohen, J., Dao, L., Morrison, P., Roth, K., Bengio, Y., Shen, B., Abbasi, A., Hoshmand-Kochi, M., Ghassemi, M., Li, H., Duong, T.Q., Predicting COVID-19 Pneumonia Severity on Chest X-ray with Deep Learning. Cureus, 12, 7, 2020, 10.7759/cureus.9448.
24. Miao, K.H. and Miao, J.H., Coronary Heart Disease Diagnosis using Deep Neural Networks. Int. J. Adv. Comput. Sci. Appl., 9, 10, 1–8, 2018.
25. Khan, S., Islam, N., Jan, Z., Ud Din, I., Rodrigues, J., A novel deep learning based framework for the detection and classification of breast cancer using transfer learning. Pattern Recognit. Lett., 125, 1–6, 2019, 10.1016/j. patrec.2019.03.022.
26. Lakshmanaprabu, S., Mohanty, S., Shankar, K., Arunkumar, N., Ramirez, G., Optimal deep learning model for classification of lung cancer on CT images. Future Gener. Comput. Syst., 92, 374–382, 2019, 10.1016/j.future.2018.10.009.
27. Laukamp, K., Thiele, F., Shakirin, G., Fully automated detection and segmentation of meningiomas using deep learning on routine multiparametric MRI. Eur. Radiol., 29, 1, 124–132, 2019.
28. Liao, F., Liang, M., Li, Z., Hu, X., Song, S., Evaluate the Malignancy of Pulmonary Nodules Using the 3-D Deep Leaky Noisy-OR Network. IEEE Trans. Neural Netw. Learn. Syst., 30, 11, 3484–3495, 2019, 10.1109/TNNLS.2019.2892409.
29. Ali, L., Rahman, A., Khan, A., Zhou, M., Javeed, A., Khan., J.A., An Automated Diagnostic System for Heart Disease Prediction Based on χ2 Statistical Model and Optimally Configured Deep Neural Network. IEEE Access, 7, 34938–34945, 2019, 10.1109/ACCESS.2019.2904800.
30. Ali, L., Niamat, A., Khan, J.A., Golilarz, N.A., Xingzhong, X., Noor, A., Nour, R., Ahmad C. Bukhari, S., An Optimized Stacked Support Vector Machines Based Expert System for the Effective Prediction of Heart Failure. 7, 54007– 54014, 2019.
31. Frid-Adar, M., Diamant, I., Klang, E., Amitai, M., Goldberger, J., Greenspan, H., GAN-based synthetic medical image augmentation for increased CNN performance in liver lesion classification. Neurocomputing, 321, 321–331, 2018, https://doi.org/10.1016/j.neucom.2018.09.013.
32. Nakrani, M.G., Sable, G.S., Shinde, U.B., Classification of COVID-19 from Chest Radiography Images Using Deep Convolutional Neural Network. J. Xidian Univ., 14, 565–569, 2020, https://doi.org/10.37896/jxu14.8/061.
33. Megersa, Y. and Alemu, G., Brain tumor detection and segmentation using hybrid intelligent algorithms. IEEE AFRICON Conference, 2015, 2015, 10.1109/AFRCON.2015.7331938.
34. Khan, M.A., An IoT Framework for Heart Disease Prediction Based on MDCNN Classifier. IEEE Access, 8, 34717–34727, 2020, 10.1109/ACCESS.2020.2974687.
35. Qjidaa, M., Mechbal, Y., Ben-fares, A., Amakdouf, H., Maaroufi, M., Alami, B., Qjidaa, H., Early detection of COVID19 by deep learning transfer Model for populations in isolated rural areas. ISCV, vol. 1, pp. 1–5, 2020, 10.1109/ISCV49265.2020.9204099.
36. Farooq, M. and Hafeez, A., COVID-ResNet: A deep learning framework for screening of COVID19 from radiographs. ArXiv, abs/2003.14395. https://arxiv.org/abs/2003.14395, 2020.
37. Imad, M., Khan, N., Ullah, F., Hassan, M.A., Hussain, A., Faiza, COVID-19 Classification based on Chest X-Ray Images Using Machine Learning Techniques. J. Comput. Sci. Technol. Stud., 2, 1–11, 2020, https://al-kindip-ublisher.com/index.php/jcsts/article/view/531.
38. Razzak, M.I., Naz, S., Zaib, A., Deep Learning for Medical Image Processing: Overview, Challenges and the Future, in: Classification in BioApps. Lecture Notes in Computational Vision and Biomechanics, vol. 26, 2017, https://doi.org/10.1007/978-3-319-65981-7_12.
39. Nibali, A., He, Z., Wollersheim, D., Pulmonary nodule classification with deep residual networks. Int. J. Comput. Assist. Radiol. Surg., 12, 10, 1799– 1808, 2017, 10.1007/s11548-017-1605-6.
40. Nithya, R. and Santhi, B., Mammogram classification using maximum difference feature selection method. J. Theor. Appl. Inf. Technol., 33, 2, 197–204, 2011.
41. Nithya, R. and Santhi, B., Breast cancer diagnosis in digital mammogram using statistical features and neural network. Res. J. Appl. Sci. Eng. Tech., 4, 24, 5480–5483, 2012.
42. Nithya, R. and Santhi, B., Mammogram analysis based on pixel intensity mean features. J. Comput. Sci., 8, 3, 329–332, 2012.
43. Nithya, R. and Santhi, B., Application of texture analysis method for mammogram density classification. J. Instrum., 12, 7, 2017.
44. Nithya, R. and Santhi, B., Computer-aided diagnosis system for mammogram density measure and classification. Biomed. Res., 28, 6, 2427–2431, 2017.
45. Lih, O.S., Jahmunah, V., San, T.R., Ciaccio, E.J., Yamakawa, T., Tanabe, M., Kobayashi, M., Faust, O., Rajendra Acharya, U., Comprehensive electrocar-diographic diagnosis based on deep learning. Artif. Intell. Med., 103, 2020, https://doi.org/10.1016/j.artmed.2019.101789.
46. Oyelade, O.N. and Ezugwu, A.E., Deep Learning Model for Improving the Characterization of Coronavirus on Chest X-ray Images Using CNN, medRXiv, 2020, https://doi.org/10.1101/2020.10.30.20222786.
47. Yildirim, O., Plawiak, P., Tan, R.-S., Rajendra Acharya, U., Arrhythmia detection using deep convolutional neural network with long duration ECG signals. Comput. Biol. Med., 102, 1, 411–420, 2018, https://doi.org/10.1016/j. compbiomed.2018.09.009.
48. Yildirim, O., Baloglu, U.B., Tan, R.-S., Ciaccio, E.J., Acharya, U.R., A new approach for arrhythmia classification using deep coded features and LSTM networks. Comput. Methods Programs Biomed., 176, 121–133, 2019, https://doi.org/10.1016/j.cmpb.2019.05.004.
49. Prakash, V. and Manjunathachari, K., Detection of brain tumour using segmentation. Int. J. Eng. Technol., 7, 688, 2018, 10.14419/ijet.v7i2.8.10559.
50. Jain, R., Gupta, M., Taneja, S., Jude Hemanth, D., Deep learning based detection and analysis of COVID-19 on chest X-ray images. Appl. Intell., 51, 1690– 1700, 2020, 10.1007/s10489-020-01902-1.
51. Rajasekar, B., Brain tumour segmentation using CNN and WT. Res. J. Pharm. Technol., 12, 10, 4613–4617, 2019, 10.5958/0974-360X.2019.00793.5.
52. Acharya, R., Fujita, H., Oh, S.L., Hagiwara, Y., Tan, J.H., Adam, M., Application of deep convolutional neural network for automated detection of myocardial infarction using ECG signals. Inf. Sci., 415–416, 190–198, 2017, https://doi.org/10.1016/j.ins.2017.06.027.
53. Rao, S. and Lingappa, B., Image analysis for MRI based brain tumour detection using hybrid segmentation and deep learning classification technique. Int. J. Intell. Eng. Syst., 12, 5, 53–62, 2019.
54. Misra, S., Jeon, S., Lee, S., Managuli, R., Jang, I.-S., Kim, C., Multi-channel transfer learning of chest x-ray images for screening of covid-19. Electronics, 9,9, 1388, 2020, https://doi.org/10.3390/electronics9091388.
55. Pathari, S. and Rahul, U., Automatic detection of COVID-19 and pneumonia from Chest X-ray using transfer learning, MedRXiv, 2020, https://doi.org/10.1101/2020.05.27.20100297.
56. Mambou, S.J., Maresova, P., Krejcar, O., Selamat, A., Kuca, K., Breast Cancer Detection Using Infrared Thermal Imaging and a Deep Learning Model. Sensors(Basel), 18, 9, 2799, 2018, 10.3390/s18092799.
57. Selvy, P.T., Dharani, V.P., Indhuja, A., Brain Tumour Detection Using Deep Learning Techniques. Int. J. Sci. Res. Comput. Sci. Eng. Inf. Technol., 169, 175, 2019, 10.32628/cseit195233.
58. SIddiqui, S.Y., Abbas, S., Khan, M.A., Naseer, I., Masood, T., Khan, K.M., Al Ghamdi, M.A., Almotiri, S.H., Intelligent decision support system for COVID-19 empowered with deep learning. Comput. Mater. Continua, 66, 1719–1732, 2021.
59. Rajpal, S., Kumar, N., Rajpal, A., COV-ELM classifier: An Extreme Learning Machine based identification of COVID-19 using Chest X-Ray Images. Image Video Process., 2020.
60. Shen, L., Margolies, L., Rothstein, J., Fluder, E., McBride, R., Sieh, W., Deep Learning to Improve Breast Cancer Detection on Screening Mammography. Sci. Rep., 9, 1, 2019, 10.1038/s41598-019-48995-4.
61. Shen, S., Han, S., Aberle, D., Bui, A., Hsu, W., An interpretable deep hierarchical semantic convolutional neural network for lung nodule malignancy classification. Expert Syst. Appl., 128, 84–95, 2019, 10.1016/j.eswa.2019.01.048.
62. Shen, W., Zhou, M., Yang, F., Learning from experts: Developing transferable deep features for patient-level lung cancer prediction. Lect. Notes Comput. Sci. (including subseries Lecture Notes in Artifical Intelligence and Lecture Notes in Bioinformatics), 9901, 124–131, 2016, 10.1007/978-3-319-46723-8_15.
63. Minaee, S., Kafieh, R., Sonka, M., Yazdani, S., Soufi, G.J., Deep-COVID: Predicting COVID-19 from chest X-ray images using deep transfer learning. Med. Image Anal., 65, 2020, https://doi.org/10.1016/j.media.2020.101794.
64. Tuli, S., Basumatary, N., Gill, S.S., Kahani, M., Arya, R.C., Wander, G.S., Buyya, R., HealthFog: An ensemble deep learning based Smart Healthcare System for Automatic Diagnosis of Heart Diseases in integrated IoT and fog computing environments. Future Gener. Comput. Syst., 104, 187–200, 2020, https://doi.org/10.1016/j.future.2019.10.043.
65. Chen, S., Ma, K., Zheng, Y., Med3D: Transfer Learning for 3D Medical Image Analysis. Comput. Vis. Pattern Recognit., 2019.
66. Skourt, A.B., Hassani, E.A., Majda, A., Lung CT image segmentation using deep neural networks. Proc. Comput. Sci., 127, 109–113, 2018, 10.1016/j. procs.2018.01.104.
67. Song, Q., Zhao, L., Luo, X., Using Deep Learning for Classification of Lung Nodules on Computed Tomography Images. J. Healthcare Eng., 2017, 7, 2017, 10.1155/2017/8314740.
68. Kim, S., An, S., Chikontwe, P., Park, S.H., Bidirectional RNN-based Few Shot Learning for 3D Medical Image Segmentation. Comput. Vis. Pattern Recognit., 2020.
69. Sriramakrishnan, P., Kalaiselvi, T., Rajeshwaran, R., Modified local ternary patterns technique for brain tumour segmentation and volume estimation from MRI multi-sequence scans with GPU CUDA machine. Biocybern. Biomed. Eng., 39, 2, 470–487, 2019, 10.1016/j.bbe.2019.02.002.
70. Thepade, S.D., Bang, S.V., Chaudhari, P.R., Dindorkar, M.R., Covid19 Identification from Chest X-ray Images using Machine Learning Classifiers with GLCM Features. ELCVIA: Electron. Lett. Comput. Vis. Image Anal., 19, 85–97, 2020, https://www.raco.cat/index.php/ELCVIA/article/view/375822.
71. Suhartono, Nguyen, P., Shankar, K., Hashim, W., Maseleno, A., Brain tumor segmentation and classification using KNN algorithm. Int. J. Eng. Adv. Technol., 8, 6, 706–711, 2019, 10.35940/ijeat.F1137.0886S19.
72. Bhattacharya. S., Reddy Maddikunta, P.K., Pham, Q.V., Gadekallu, T.R., Krishnan S.S.R., Chowdhary, C.L., Alazab, M., Jalil Piran, M., Deep learning and medical image processing for coronavirus (COVID-19) pandemic: A survey. Sustain. Cities Soc.,2021.
73. Tan, T., Das, B., Soni, R., Fejes, M., Ranjan, S., Szabo, D.A., Melapudi, V., Shriram, K.S., Agrawal, U., Rusko, L., Herczeg, Z., Darazs, B., Tegzes, P., Ferenczi, L., Mullick, R., Avinash, G., Pristine annotations-based multi-modal trained artificial intelligence solution totriage chest x-ray for COVID19, arXiv, 2020, https://arxiv.org/abs/2011.05186.
74. Wang, H., Zhou, Z., Li, Y. et al., Comparison of machine learning methods for classifying mediastinal lymph node metastasis of non-small cell lung cancer from 18F-FDG PET/CT images. EJNMMI Res., 7, 11, 2017. https://doi.org/10.1186/s13550-017-0260-9
75. Weiss, K., Khoshgoftaar, T.M., Wang, D., A survey of transfer learning. J. Big Data, 3, 9, 2016, 10.1186/s40537-016-0043-6.
76. Wu, W., Chen, A., Zhao, L., Corso, J., Brain tumor detection and segmentation in a CRF (conditional random fields) framework with pixel-pairwise affinity and superpixel-level features. Int. J. Comput. Assist. Radiol. Surg., 9, 2, 241–253, 2014, 10.1007/s11548-013-0922-7.
77. Fan, X., Yao, Q., Cai, Y., Miao, F., Sun, F., Li, Y., Multiscaled Fusion of Deep Convolutional Neural Networks for Screening Atrial Fibrillation From Single Lead Short ECG Recordings. IEEE J. Biomed. Health Inform., 22, 1744–1753, 2018.
78. Xu, X., Jiang, X., Ma, C., Du, P., Li, X., Lv, S., Yu, L., Ni, Q., Chen, Y., Su, J., Lang, G., Li, Y., Zhao, H., Liu, J., Xu, K., Ruan, L., Sheng, J., Qiu, Y., Wu, W., Li, L., A Deep Learning System to Screen Novel Coronavirus Disease 2019 Pneumonia. Engineering, 6, 10, 1122–1129, 2020, https://doi.org/10.1016/j.eng.2020.04.010.
79. Xie, Y., Xia, Y., Zhang, J., Song, Y., Feng, D., Fulham, M., Cai, W., Knowledge-based Collaborative Deep Learning for Benign-Malignant Lung Nodule Classification on Chest CT. IEEE Trans. Med. Imaging, 38, 4, 991–1004,, 2019, 10.1109/TMI.2018.2876510.
80. Zhao, X., Liu, L., Qi, S., Teng, Y., Li, J., Qian, W., Agile convolutional neural network for pulmonary nodule classification using CT images. Int. J. Comput. Assist. Radiol. Surg., 13, 4, 585–595, 2018, 10.1007/s11548-017-1696-0.
- *Corresponding author: [email protected]
