
444 Handb ook of Big Data
Alternatively, one might define Ψ
C
(P
0
) as the average over all genes j in cluster C of the
effect of treatment A on Y (j), controlling for W , defined as
Ψ
C
(P
0
)=
1
| C |
|C|
j=1
E
0
{E
0
(Y (j) | A =1,W) − E
0
(Y (j) | A =0,W)}
Let
ˆ
Ψ
C
: M
NP
→ IR be an estimator of Ψ
C
(P
0
), for instance, inverse probability
of treatment weighted (IPTW [12]), or targeted maximum likelihood estimator (TMLE
[11,13]). If one assumes that the regularity conditions hold so that, for instance, the TMLE
estimator,
ˆ
Ψ
C
(P
n
), is asymptotically linear with the influence curve IC
C
(P
0
), then
ˆ
Ψ
C
(P
n
) − Ψ
C
(P
0
)=(P
n
− P
0
)IC
C
(P
0
)+R
C,n
where R
C,n
= o
P
(1/
√
n). We define Ψ
P
n,v
c
: M→IR a s Ψ
P
n,v
c
=Ψ
ˆ
C
(P
n,v
c
)
,thatis,the
causal effect of treatment on the data-adaptively determined cluster
ˆ
C(P
n,v
c
). Similarly, we
define
ˆ
Ψ
P
n,v
c
: M
NP
→ IR a s
ˆ
Ψ
P
n,v
c
=
ˆ
Ψ
ˆ
C(P
n,v
c
)
, that is, the TMLE of the W -controlled
effect of treatment of this data-adaptively determined cluster, treating the latter as given.
The estimand of interest is thus defined as ψ
n,0
=Ave{Ψ
P
n,v
c
(P
0
)} and its estimator is
ψ
n
=Ave{
ˆ
Ψ
P
n,v
c
(P
n,v
)}. Thus, for a given split, we use the parameter-generating sample
P
n,v
c
to generate a cluster
ˆ
C(P
n,v
c
) and the corresponding TMLE of
ˆ
Ψ
ˆ
C
(P
n,v
c
)
(P
0
) applied
to the estimation sample P
n,v
, and these sample-split specific estimators are averaged across
the V sample splits. By assumption we have for each split v:
ˆ
Ψ
ˆ
C(P
n,v
c
)
(P
n,v
) − Ψ
ˆ
C(P
n,v
c
)
(P
0
)=(P
n,v
− P
0
)IC
ˆ
C(P
n,v
c
)
(P
0
)+R
ˆ
C(P
n,v
c
),n
where we now assume that (unconditionally) R
ˆ
C
(P
n,v
c
),n
= o
P
(1/
√
n). In addition, we
assume that P
0
{IC
ˆ
C(P
n,v
c
)
(P
0
)}
2
converges to P
0
{IC
ˆ
C(P
0
)
(P
0
)}
2
for a limit cluster
ˆ
C(P
0
).
Application of Theorem 24.1 now proves that ψ
n
− ψ
n,0
is asymptotically linear with the
influence curve IC
ˆ
C(P
0
)
(P
0
) so that it is asymptotically normally distributed with mean
zero and variance σ
2
= P
0
IC
ˆ
C(P
0
)
(P
0
)
2
.
There are many examples of potential applications of this approach too numerous to list
(see [2] for more examples and extensive simulations). However, one particularly important
one is the estimation of the causal effect of particular treatment rules that are learned from
data. This one has obvious practical implications in all sorts of fields, including medicine,
political science, and any discipline where of interest is the impact of an approach that
tailors an intervention to the characteristics of the statistical units, and the data must be
used to learn the best approach.
24.4 Data Analysis: ACIT Variable Importance Trauma Study
We revisit estimating VIMs of the form 24.2 for the ACIT trauma data. The target
population are those patients that survive up to 6 hours after their injury, and the outcome
Y is the indicator of death from 6 to 24 hours, where, among the n = 1277 observations
(subjects) alive at 6 hours, the proportion of deaths in this next interval is 3.5%. We
estimate the variable importance from a combination of ordered continuous variables,
ordered discrete, and factors for a total of 108 potential predictors. Each of these 108
variables is treated as the variable of interest, A, and the remainder as covariates. This
Mining with Inference 445
is an extremely messy data (missing values, sparsity, etc.), and thus, for each variable
importance we undergo a number of automated data-processing steps to make estimation
viable. These include:
1. Drop variables missing for more than 50% of observations.
2. Automatically determine which variables are ordered and unordered factors.
3. Drop observations variables for which there distribution is too uneven so that
there is not enough experimentation across different levels to estimate a VIM.
Then, for each variable (when current variable of interest), we performed the following steps.
1. For a continuous variable, we constructed new ordered discrete variables (integers)
that map the original value into intervals defined by the empirical deciles of the
variable. This results in new ordered variables with values 1, ..., max(A), where
max(A) = 10 unless the original variable has fewer unique values. We then further
lump these values into groups by histogram density estimation using the penalized
likelihood approach [14,15] to bin values, avoiding very small cell sizes in the
distribution A.
2. For each variable, we generated basis functions that indicate which observations
have missing values, so that for each original variable, say W
j
, there is a new basis
(Δ
j
, Δ
j
∗W
j
), where Δ
j
=1ifW
j
is not missing for an observation, 0 otherwise
(see below for how this is used in the algorithm).
3. Use hierarchical clustering routine HOPACH [16] to cluster adjustment variables
(the matrices of all other predictors besides the current A as well as the matrix of
associated missingness indicators) to reduce the dimension of the adjustment set.
Each of these processing steps is tunable depending on sample size to the point where the
data are very close to the original form. These steps allow a very general messy dataset
of mixed variable types that can be automatically processed for VIM analysis. Thus, after
processing, for each VIM analysis we can represent the data as i.i.d., O =(W, A, Y ) ∼ P
0
∈
M, W being all other covariates besides the current A as well as the basis functions related
to missingness observations for covariates.
We estimate our data-adaptive, VIM based on algorithms motivated by both theorems
above. Specifically, we use the approach of Theorem 24.1 by performing sample splitting
using two-fold cross-validation (splitting randomly in equal samples). The parameter is first
defined by using the training sample to define a
L
(P
n,v
c
). To do so, we estimate, for each of
values, a, of the discretized A ∈A, an estimand motivated by the causal parameter E(Y
a
).
Under identifiability assumptions (e.g., randomization, positivity; see Chapter 4 in [11]), we
can identify E(Y
a
)as
E
0
{E
0
(Y | A = a, W )} = E
0
{Q
0
(a, W )} (24.6)
where 0 indicated under P
0
,andQ
0
(a, W ) is the true regression (conditional mean) function
of Y on (A, W ). If we knew Q
0
, we would know the true levels of A to compare, that is,
a
L
≡ argmin
a∈A
E
0
{Q
0
(a, W)}
a
H
≡ argmax
a∈A
E
0
{Q
0
(a, W)}
so we could then estimate Equation 24.1 without having to discover values of (a
L
,a
H
)using
the data. However, we do not know Q
0
and must estimate it to estimate Equation 24.6 to
empirically define (a
L
,a
H
). In the context of Theorem 24.1, we then use the training sample
to define these levels. We do so using the following algorithm:
446 Handb ook of Big Data
1. Estimate θ
0
(a) ≡ E
0,W
{Q
0
(a, W )} for all levels a ∈Ausing a semi-parametric,
locally efficient data-adaptive method TMLE [11].
a. To do so requires an initial estimate of the regression Q
0
(A, W ) and we use
the SuperLearner algorithm [17], an ensemble learner that itself uses cross-
validation to derive an optimal weighted combination of selected learners. For
this we used (1) LASSO via the glmnet package in R [18], (2) ADD OTHERS.
b. This also requires an estimate of the so-called treatment mechanism
(g
0
(a; W ) ≡ P
0
(A = a | W )) and the glmnet package was also used for this.
We can define these estimates for a specific training sample v
c
as
ˆ
θ
P
n,v
c
(a).
2. Select the levels of A to compare in the corresponding training sample as
a
L
(P
n,v
c
) = argmin
a∈A
ˆ
θ
P
n,v
c
(a)
a
H
(P
n,v
c
) = argmax
a∈A
ˆ
θ
P
n,v
c
(a)
3. On the corresponding estimation sample, estimate the parameter
Ψ
P
n,v
c
(P
0
) ≡ θ
0
(a
H
(P
n,v
c
)) − θ
0
(a
L
(P
n,v
c
)) (24.7)
We estimated Ψ
P
n,v
c
(P
0
) on the corresponding estimation sample (say
ˆ
Ψ
P
n,v
c
(P
n,v
)) using the same combination of SuperLearner and TMLE described
above as used for defining (a
L
(P
n,v
c
),a
H
(P
n,v
c
)).
4. Derive the influence curve of these estimators on the validation sample. In this
case, the estimated influence curve is
IC
P
n,v
c
(P
n,v
)=Y −
I{A=a
H
(P
n,v
c
)}
g
P
n,v
(a
H
(P
n,v
c
);W )
−
I{A=a
L
(P
n,v
c
)}
g
P
n,v
(a
L
(P
n,v
c
);W )
{Y −Q
P
n,v
(A, W )}
+
Q
P
n,v
(a
H
(P
n,v
c
),W) −Q
P
n,v
(a
L
(P
n,v
c
),W)
− Ψ
P
n,v
c
(P
n,v
)
where Q
P
n,v
and g
P
n,v
indicate that these functions were estimated on the
estimation sample P
n,v
.
5. Derive the estimated sample variance of the influence curve: σ
2
n,v
≡ P
n,v
IC
P
n,v
c
(P
n,v
)
2
.
6. Repeat steps 1–5 above for every combination of estimation and validation
sample to get the entire set of parameter and variance estimates, (
ˆ
Ψ
P
n,v
c
(P
n,v
),
σ
2
P
n,v
),v =1,...,V.
7. Average the estimates to get the estimate (Equation 24.4) of the average
parameter: ψ
n
=Ave{
ˆ
Ψ
P
n,v
c
(P
n,v
)}, as well as the average estimated variances,
σ
2
n
=Ave{σ
2
n,v
}.
8. Derive confidence intervals, and p-values based using quantities calculated in 7.
9. Repeat steps 1–8 for every variable to consider as an A by switching places with
the current A and one of the W .
10. Basedonthep-value of the test of null: H
0
: ψ
n,0
≤ 0, adjust for multiple
comparisons by controlling the false discovery rate (FDR; [19])
The results are an ordered list (by statistical significance) of the VIM (the ψ
n
), statistical
inference adjusted for multiple comparisons, along with information about the estimates and
the levels of (a
L
(P
n,v
c
),a
H
(P
n,v
c
)) chosen for each combination of training and estimation
samples.
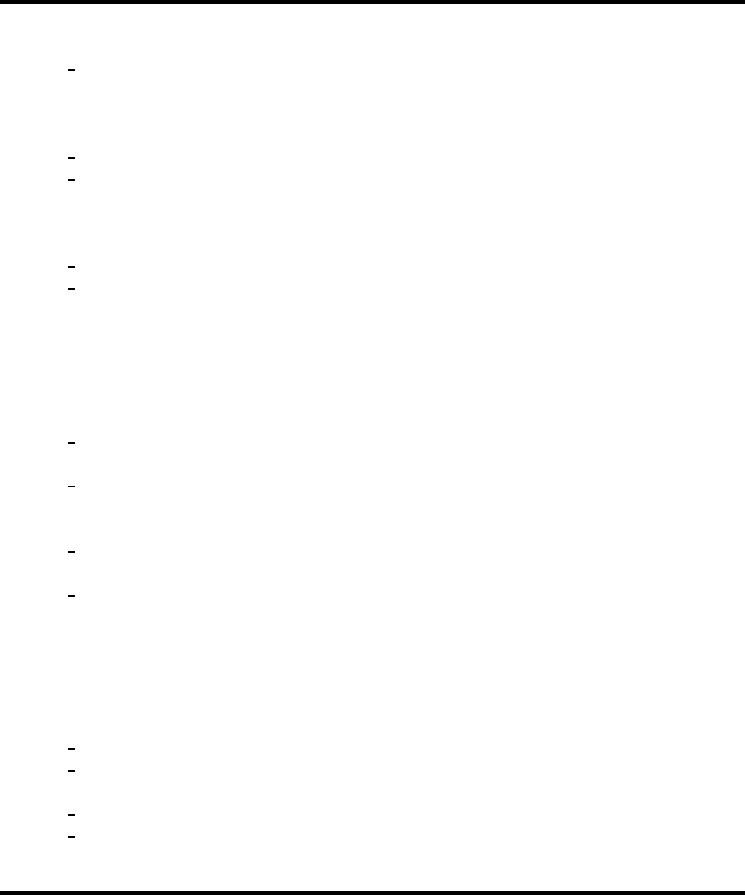
Mining with Inference 447
24.4.1 Results
The names of the potential predictors along with the type of variable (ordered or unordered
factor) are listed in Table 24.1. This table is ordered by the p-value of the test: H
0
: ψ
n
≤ 0,
so most significant estimates at top. There were 108 original variables examined, but only
72 with sufficient data to estimate a VIM. Table 24.2 has the estimation sample-specific
results for the same ordered list. The definition of the (a
L
(P
n,v
c
),a
H
(P
n,v
c
)) for the two
training samples is related to the original discretization, so, for instance, if it states that
a
L
(P
n,v
c
)is(x, 1], then a
L
(P
n,v
c
) is the indicator of being in the lowest decile. One can
see some shortcomings of the data from these results. The outcome is rare enough, that
TABLE 24.1
ACIT variables and description—note variables are ordered by statistical significance.
Name Var. type Description
tbi Factor Traumatic brain injury
hr6
basedefexc Ordered Hour 6 base deficit/excess
ortho Factor Orthopedic injury
alcabuse Factor Alcohol use
race Factor Race
hr0
factorviii Ordered Hour 0 factor VIII
hr0
ptt Ordered Hour 0 partial thromboplastin time
heightcm Ordered Height
edworsttemp Ordered ED worst temp
aisface2 Ordered Abbreviated injury scale: face
hr0
factorv Ordered Hour 0 factor V
hr0
atiii Ordered Hour 0 antithrombin III
aisextremity5 Ordered Abbreviated injury scale: extremity
male Factor Gender male?
edlowestsbp Ordered ED lowest SBP
latino Factor Latino
pbw Ordered Predicted body weight
edlowesthr Ordered ED lowest HR
hr0
temp Ordered Hour 0 temperature
blunt Factor Mechanism of injury—blunt?
hr0
map Ordered Hour 0 mean arterial pressure
aisabdomen4 Ordered Abbreviated injury scale: abdomen
iss Ordered Injury severity score
hr0
resprate Ordered Hour 0 respiratory rate
numribfxs Ordered Number of rib fractures
hr0
factorx Ordered Hour 0 factor X
patientbloodtype Factor Patient blood type
edadmittemp Ordered ED admit temp
edhighestsbp Ordered ED highest SBP
age Ordered AGE at time of injury
insurancesource Factor Insurance source
mechtype Factor Mechanism type
hr0
basedefexc Ordered Hour 0 base deficit/excess
hr0
ph Ordered Hour 0 pH
ali Factor Acute lung injury
hr0
pc Ordered Hour 0 protein C
hr0
factorix Ordered Hour 0 factor IX
edworstrr Ordered ED worst RR
aischest3 Ordered Abbreviated injury scale: chest
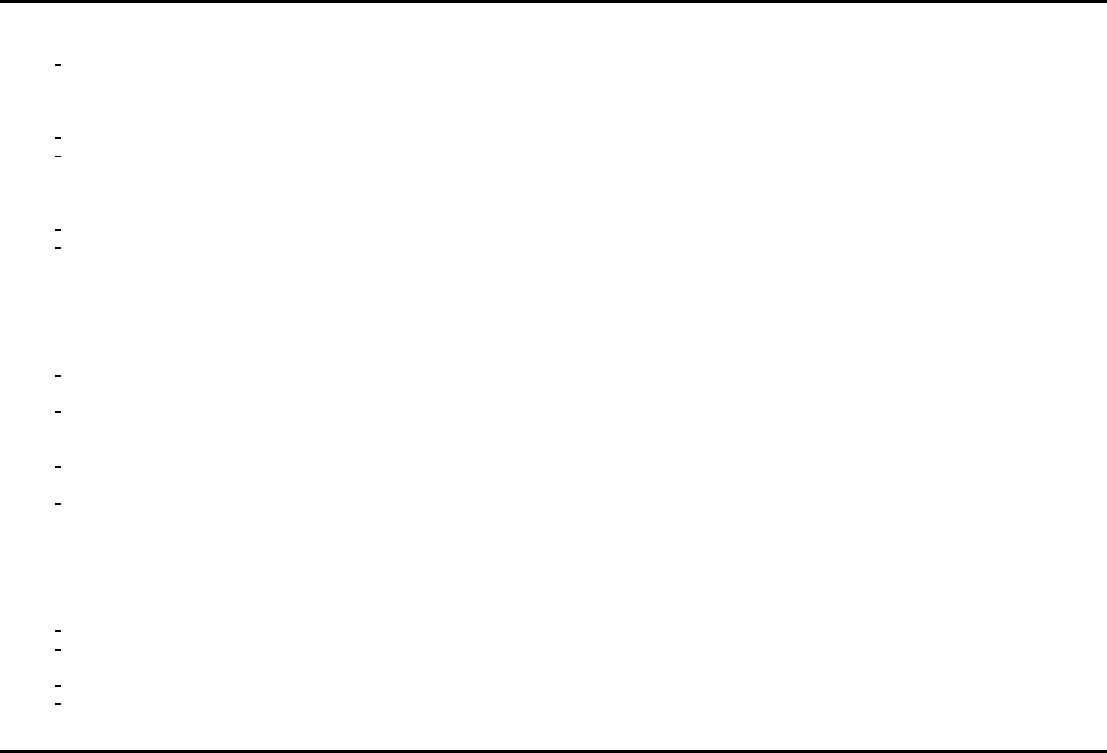
448 Handb ook of Big Data
TABLE 24.2
ACIT variable importance results by the estimation sample.
Predictor
ˆ
Ψ
P
n,(v=1)
c
ˆ
Ψ
P
n,(v=2)
c
a
L
(P
n,(v=1)
c ) a
H
(P
n,(v=1)
c ) a
L
(P
n,(v=2)
c ) a
H
(P
n,(v=2)
c )
tbi 0.0636 0.1130 No Yes No Yes
hr6
basedefexc 0.1283 0.2141 (1,10.1] (0.95,1] (1,10] (0.9,1]
ortho 0.0300 0.0554 Yes No Yes No
alcabuse 0.0565 0.0588 No Unknown No Unknown
race 0.0961 0.0407 White Asian White Asian
hr0
factorviii 0.1753 0.0947 (1,10] (0.9,1] (1,10.1] (0.9,1]
hr0
ptt 0.0895 0.0431 (1,9] (9,10] (0.9,6] (6,10]
heightcm 0.0800 0.0156 (1,10] (0.9,1] (1,10.1] (0.9,1]
edworsttemp 0.0506 0.0582 (1,10.1] (0.95,1] (3,10.1] (0.9,1]
aisface2 0.0085 0.0266 (1,4] (0.97,1] (1,4] (0.97,1]
hr0
factorv 0.0166 0.059 (1,10] (0.9,1] (1,10.1] (0.9,1]
hr0
atiii 0.0159 0.0322 (1,10] (0.9,1] (1,10] (0.9,1]
aisextremity5 0.0164 0.0189 (1,5] (0.96,1] (1,5] (0.96,1]
male 0.0648 −0.0159 Male Female Male Female
edlowestsbp 0.0334 0.0311 (1,10] (0.9,1] (1,10] (0.9,1]
latino 0.019 0.0154 Yes No Yes No
pbw 0.0577 0.0107 (1,10] (0.9,1] (1,10.1] (0.9,1]
edlowesthr 0.0317 0.0557 (1,10.1] (0.9,1] (1,10] (0.9,1]
hr0
temp 0.029 0.0401 (1,10.1] (0.9,1] (1,10.1] (0.9,1]
blunt 0.0293 −0.0018 Penetrating Blunt Penetrating Blun t
hr0
map 0.0506 0.0053 (0.9,1] (9,10] (0.9,1] (1,10]
aisabdomen4 0.0159 0.0059 (1,4] (0.97,1] (1,4] (0.97,1]
iss 0.0329 0.0634 (0.9,5] (5,9] (1,7] ( 7,9]
hr0
resprate −0.0059 0.0268 (1,10] (0.9,1] (2,10] (1,2]
numribfxs 0.0179 0.0103 (1,4] ( 0.97,1] (1,4] (0.97,1]
hr0
factorx 0.0461 −0.0087 (1,10] (0.9,1] (6,10] (0.9,1]
patientbloodtype 0.0219 0.0298 A+ A− A+ A−
edadmittemp −0.0008 0.0220 (1,10.1] (0.9,1] (3,10.1] (1,3]
edhighestsbp 0.0120 0.0150 (1,9] (9,10] (1,10] (0.9,1]
age −0.0095 0.0280 (1,10] (0.9,1] (1,9] (9,10]
insurancesource 0.0235 −0.0124 No insurance Medical No insurance Medicare
mechtype 0.0801 −0.0328 PVA Found down Found down PVA
hr0
basedefexc 0.0083 0.0095 (1,10.1] (0.9,1] (1,10] (0.9,1]
hr0
ph 0.0509 −0.0316 (1,10] (0.9,1] (0.9,1] (1,10]
ali −0.0275 0.0116 No Yes Yes No
hr0
pc 0.0058 −0.0477 (1,10] (0.9,1] (0.9,1] (1,10]
hr0
factorix −0.0293 −0.0192 (1,10] (0.9,1] (0.9,1] (1,10]
edworstrr −0.0106 − 0.0644 (1,9.1] (0.9,1] (0.9,1] (1,9]
aischest3 −0.s0270 −0.0326 (1,5.1] (0.97,1] (0.96,1] (1,5]
..................Content has been hidden....................
You can't read the all page of ebook, please click here login for view all page.
