Each year, many people fall ill and sometimes even die from ingesting poisonous wild mushrooms. Since many mushrooms are very similar to each other in appearance, occasionally even experienced mushroom gatherers are poisoned.
Unlike the identification of harmful plants such as a poison oak or poison ivy, there are no clear rules such as "leaves of three, let them be" to identify whether a wild mushroom is poisonous or edible. Complicating matters, many traditional rules, such as "poisonous mushrooms are brightly colored," provide dangerous or misleading information. If simple, clear, and consistent rules were available to identify poisonous mushrooms, they could save the lives of foragers.
Because one of the strengths of rule learning algorithms is the fact that they generate easy-to-understand rules, they seem like an appropriate fit for this classification task. However, the rules will only be as useful as they are accurate.
To identify rules for distinguishing poisonous mushrooms, we will utilize the Mushroom dataset by Jeff Schlimmer of Carnegie Mellon University. The raw dataset is available freely at the UCI Machine Learning Repository (http://archive.ics.uci.edu/ml).
The dataset includes information on 8,124 mushroom samples from 23 species of gilled mushrooms listed in Audubon Society Field Guide to North American Mushrooms (1981). In the Field Guide, each of the mushroom species is identified "definitely edible," "definitely poisonous," or "likely poisonous, and not recommended to be eaten." For the purposes of this dataset, the latter group was combined with the "definitely poisonous" group to make two classes: poisonous and nonpoisonous. The data dictionary available on the UCI website describes the 22 features of the mushroom samples, including characteristics such as cap shape, cap color, odor, gill size and color, stalk shape, and habitat.
We begin by using read.csv(), to import the data for our analysis. Since all the 22 features and the target class are nominal, in this case, we will set stringsAsFactors = TRUE and take advantage of the automatic factor conversion:
> mushrooms <- read.csv("mushrooms.csv", stringsAsFactors = TRUE)
The output of the str(mushrooms) command notes that the data contain 8,124 observations of 23 variables as the data dictionary had described. While most of the str() output is unremarkable, one feature is worth mentioning. Do you notice anything peculiar about the veil_type variable in the following line?
$ veil_type : Factor w/ 1 level "partial": 1 1 1 1 1 1 ...
If you think it is odd that a factor has only one level, you are correct. The data dictionary lists two levels for this feature: partial and universal. However, all the examples in our data are classified as partial. It is likely that this data element was somehow coded incorrectly. In any case, since the veil type does not vary across samples, it does not provide any useful information for prediction. We will drop this variable from our analysis using the following command:
> mushrooms$veil_type <- NULL
By assigning NULL to the veil type vector, R eliminates the feature from the mushrooms data frame.
Before going much further, we should take a quick look at the distribution of the mushroom type class variable in our dataset:
> table(mushrooms$type) edible poisonous 4208 3916
About 52 percent of the mushroom samples (N = 4,208) are edible, while 48 percent (N = 3,916) are poisonous.
For the purposes of this experiment, we will consider the 8,214 samples in the mushroom data to be an exhaustive set of all the possible wild mushrooms. This is an important assumption, because it means that we do not need to hold some samples out of the training data for testing purposes. We are not trying to develop rules that cover unforeseen types of mushrooms; we are merely trying to find rules that accurately depict the complete set of known mushroom types. Therefore, we can build and test the model on the same data.
If we trained a hypothetical ZeroR classifier on this data, what would it predict? Since ZeroR ignores all of the features and simply predicts the target's mode, in plain language, its rule would state that all the mushrooms are edible. Obviously, this is not a very helpful classifier, because it would leave a mushroom gatherer sick or dead with nearly half of the mushroom samples bearing the possibility of being poisonous. Our rules will need to do much better than this in order to provide safe advice that can be published. At the same time, we need simple rules that are easy to remember.
Since simple rules can often be extremely predictive, let's see how a very simple rule learner performs on the mushroom data. Toward the end, we will apply the 1R classifier, which will identify the most predictive single feature of the target class and use it to construct a set of rules.
We will use the 1R implementation in the RWeka package called OneR(). You may recall that we had installed RWeka in Chapter 1, Introducing Machine Learning, as a part of the tutorial on installing and loading packages. If you haven't installed the package per these instructions, you will need to use the install.packages("RWeka") command and have Java installed on your system (refer to the installation instructions for more details). With these steps complete, load the package by typing library(RWeka):
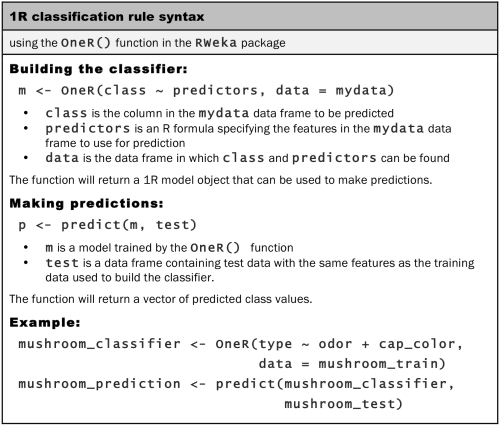
The OneR() implementation uses the R formula syntax to specify the model to be trained. The formula syntax uses the ~ operator (known as the tilde) to express the relationship between a target variable and its predictors. The class variable to be learned goes to the left of the tilde, and the predictor features are written on the right, separated by + operators. If you like to model the relationship between the y class and predictors x1 and x2, you could write the formula as y ~ x1 + x2. If you like to include all the variables in the model, the special term . can be used. For example, y ~ . specifies the relationship between y and all the other features in the dataset.
Tip
The R formula syntax is used across many R functions and offers some powerful features to describe the relationships among predictor variables. We will explore some of these features in the later chapters. However, if you're eager for a sneak peek, feel free to read the documentation using the ?formula command.
Using the type ~ . formula, we will allow our first OneR() rule learner to consider all the possible features in the mushroom data while constructing its rules to predict type:
> mushroom_1R <- OneR(type ~ ., data = mushrooms)
To examine the rules it created, we can type the name of the classifier object, in this case, mushroom_1R:
> mushroom_1R odor: almond -> edible anise -> edible creosote -> poisonous fishy -> poisonous foul -> poisonous musty -> poisonous none -> edible pungent -> poisonous spicy -> poisonous (8004/8124 instances correct)
In the first line of the output, we see that the odor feature was selected for rule generation. The categories of odor, such as almond, anise, and so on, specify rules for whether the mushroom is likely to be edible or poisonous. For instance, if the mushroom smells fishy, foul, musty, pungent, spicy, or like creosote, the mushroom is likely to be poisonous. On the other hand, mushrooms with more pleasant smells like almond and anise, and those with no smell at all are predicted to be edible. For the purposes of a field guide for mushroom gathering, these rules could be summarized in a simple rule of thumb: "if the mushroom smells unappetizing, then it is likely to be poisonous."
The last line of the output notes that the rules correctly predicted the edibility of 8,004 of the 8,124 mushroom samples or nearly 99 percent of the mushroom samples. We can obtain additional details about the classifier using the summary() function, as shown in the following example:
> summary(mushroom_1R) === Summary === Correctly Classified Instances 8004 98.5229 % Incorrectly Classified Instances 120 1.4771 % Kappa statistic 0.9704 Mean absolute error 0.0148 Root mean squared error 0.1215 Relative absolute error 2.958 % Root relative squared error 24.323 % Coverage of cases (0.95 level) 98.5229 % Mean rel. region size (0.95 level) 50 % Total Number of Instances 8124 === Confusion Matrix === a b <-- classified as 4208 0 | a = edible 120 3796 | b = poisonous
The section labeled Summary lists a number of different ways to measure the performance of our 1R classifier. We will cover many of these statistics later on in Chapter 10, Evaluating Model Performance, so we will ignore them for now.
The section labeled Confusion Matrix is similar to those used before. Here, we can see where our rules went wrong. The key is displayed on the right, with a = edible and b = poisonous. Table columns indicate the predicted class of the mushroom while the table rows separate the 4,208 edible mushrooms from the 3,916 poisonous mushrooms. Examining the table, we can see that although the 1R classifier did not classify any edible mushrooms as poisonous, it did classify 120 poisonous mushrooms as edible—which makes for an incredibly dangerous mistake!
Considering that the learner utilized only a single feature, it did reasonably well; if one avoids unappetizing smells when foraging for mushrooms, they will almost avoid a trip to the hospital. That said, close does not cut it when lives are involved, not to mention the field guide publisher might not be happy about the prospect of a lawsuit when its readers fall ill. Let's see if we can add a few more rules and develop an even better classifier.
For a more sophisticated rule learner, we will use JRip(), a Java-based implementation of the RIPPER rule learning algorithm. As with the 1R implementation we used previously, JRip() is included in the RWeka package. If you have not done so yet, be sure to load the package using the library(RWeka) command:

As shown in the syntax box, the process of training a JRip() model is very similar to how we previously trained a OneR() model. This is one of the pleasant benefits of the functions in the RWeka package; the syntax is consistent across algorithms, which makes the process of comparing a number of different models very simple.
Let's train the JRip() rule learner as we did with OneR(), allowing it to choose rules from all the available features:
> mushroom_JRip <- JRip(type ~ ., data = mushrooms)
To examine the rules, type the name of the classifier:
> mushroom_JRip JRIP rules: =========== (odor = foul) => type=poisonous (2160.0/0.0) (gill_size = narrow) and (gill_color = buff) => type=poisonous (1152.0/0.0) (gill_size = narrow) and (odor = pungent) => type=poisonous (256.0/0.0) (odor = creosote) => type=poisonous (192.0/0.0) (spore_print_color = green) => type=poisonous (72.0/0.0) (stalk_surface_below_ring = scaly) and (stalk_surface_above_ring = silky) => type=poisonous (68.0/0.0) (habitat = leaves) and (cap_color = white) => type=poisonous (8.0/0.0) (stalk_color_above_ring = yellow) => type=poisonous (8.0/0.0) => type=edible (4208.0/0.0) Number of Rules : 9
The JRip() classifier learned a total of nine rules from the mushroom data. An easy way to read these rules is to think of them as a list of if-else statements, similar to programming logic. The first three rules could be expressed as:
- If the odor is foul, then the mushroom type is poisonous
- If the gill size is narrow and the gill color is buff, then the mushroom type is poisonous
- If the gill size is narrow and the odor is pungent, then the mushroom type is poisonous
Finally, the ninth rule implies that any mushroom sample that was not covered by the preceding eight rules is edible. Following the example of our programming logic, this can be read as:
- Else, the mushroom is edible
The numbers next to each rule indicate the number of instances covered by the rule and a count of misclassified instances. Notably, there were no misclassified mushroom samples using these nine rules. As a result, the number of instances covered by the last rule is exactly equal to the number of edible mushrooms in the data (N = 4,208).
The following figure provides a rough illustration of how the rules are applied to the mushroom data. If you imagine everything within the oval as all the species of mushroom, the rule learner identified features or sets of features, which separate homogeneous segments from the larger group. First, the algorithm found a large group of poisonous mushrooms uniquely distinguished by their foul odor. Next, it found smaller and more specific groups of poisonous mushrooms. By identifying covering rules for each of the varieties of poisonous mushrooms, all of the remaining mushrooms were found to be edible. Thanks to Mother Nature, each variety of mushrooms was unique enough that the classifier was able to achieve 100 percent accuracy.


