3 Online health information semantic search and exploration: reporting on two prototypes for performing information extraction on both a hospital intranet and the world wide web
Abstract: In this chapter, we apply ontology-based information extraction to unstructured natural language sources to help enable semantic search of health information. We propose a general architecture capable of handling both private and public data. Two of our novel systems that are based on this architecture are presented here. The first system, MedInX, is a Medical Information eXtraction system which processes textual clinical discharge records, performing automatic and accurate mapping of free text reports onto a structured representation. MedInX is designed to be used by health professionals, and by hospital administrators and managers, allowing its users to search the contents of such automatically populated ontologies. The second system, SPHInX, attempts to perform semantic search on health information publicly available on the web in Portuguese. The potential of the proposed approach is clearly shown with usage examples and evaluation results.
3.1 Introduction
More and more healthcare institutions store vast amounts of information about users, procedures, and examinations, as well as the findings, test results, and diagnoses, respectively. Other institutions, such as the Government, increasingly disclose health information on varied topics of concern to the public writ large. Health research is one of the most active areas, resulting in a steady flow of publications reporting on new findings and results.
In recent years, the Internet has become one of the most important tools to obtain medical and health information. Standard web search is by far the most common interface for such information (Abraham & Reddy 2007). Several general search engines such as Google, Yahoo! and Bing currently play an important role in obtaining medical information for both medical professionals and lay persons (Wang et al. 2012). However, these general search engines do not allow the end-user to obtain a clear and organized presentation of the available health information. Instead, it is more or less of a hit or miss, random return of information on any given search. In fact, medicine-related information search is different from other information searches, since users often use medical terminology, disease knowledge, and treatment options in their search (Wang et al. 2012).
Much of the information that would be of interest to private citizens, researchers, and health professionals is found in unstructured text documents. Efficient access to this information implies the development of search systems capable of handling the technical lexicon of the domain area, entities such as drugs and exams, and the domain structure. Such search systems are said to perform semantic search as they base the search on the concepts asked and not so much on the words used in the query (Guha, McCool & Miller 2003). Semantic search maintains several advantages over search based on surface methods, such as those that directly index text words themselves rather than underlying concepts. Three main advantages of concept-based search are: (1) they usually produce smaller sets of results, as they are able to identify and remove semantically duplicated results and/or semantically irrelevant results; (2) they can integrate related information scattered across documents; frequently answers are obtained by compounding information from two or more sources; and (3) they can retrieve relevant results even when the question and answer do not have common words, since these systems can be aware of similar concepts, synonyms, meronyms, antonyms, etc.
Semantic search involves representing the concepts of a domain and the relations between them, organizing, in this way, the information according to its semantics and forming a knowledge representation of the world or some part of it. This representation is called ontology, which is formally defined as an explicit specification of a shared conceptualization (Gruber 1993). Ontology describes a hierarchy of concepts related by subsumption relationships, and can include axioms to express other relationships between concepts and to constrain their intended interpretation. The usage of ontology to explicitly define the application domain brings large benefits from the viewpoint of information accessibility, maintainability and interoperability, as it formalizes and allows the application’s view of the world to be made public (Guarino 1998). Also, with the emergence of semantic reasoners, software that is able to infer logical consequences from a set of asserted facts or axioms, it is possible to verify the coherence of the stored information and to infer new information from the contents of ontology (Sirin & Parsia 2004).
However, there is still the challenge of bridging the gap between the needed semantically structured information and the original text content. The acquisition of specific and relevant pieces of information from texts, and respective storage in a coherent framework, is called information extraction (Cowie & Lehnert 1996). The general problem of information extraction (IE) involves the analysis of natural language texts, such as English or Portuguese texts, to determine the semantic relations among the existing entities and the events they participate in, namely their relations. Natural language texts can be unstructured, plain texts, and/or semi-structured machine-readable documents, with some kind of markup. The information to be retrieved can be entities, classes of objects and events, and relationships between them. Informally, IE is the task of detecting elements such as “who” did “what” to “whom,” “when” and “where,” in unstructured free text information sources, and using those elements to populate structured information sources (Gaizauskas & Wilks 1998; Màrquez et al. 2008).
IE is different from information retrieval (IR), which is the task usually performed by current search engines such as Google and Bing. Whereas IE aims to extract relevant information from documents, IR aims to retrieve those relevant documents themselves from collections. For example, universities and other public libraries use IR systems to provide access to books, journals and other documents. However, in such cases, after querying search engines the users still have to read through those documents brought up in their search to find the information they were looking for. When the goal is to explore data, obtain a summary of facts reported in large amounts of documents or have facts presented in tables, IE becomes a much more relevant technology than IR (McNaught & Black 2006).
A typical IE system has two main subtasks: entity recognition and relation extraction. Entity recognition seeks to locate and classify atomic elements in natural language texts into predefined categories, while relation extraction tries to identify the relations between the entities in order to fill predefined templates. Two important challenges exist in IE. One arises from the variety of ways of expressing the same fact. The other challenge, shared by almost all NLP tasks, is due to the great expressiveness of natural languages, which can have ambiguous structure and meaning.
The chapter is structured as follows: the next two sections provide background information and an overview of related work about information search in general and in the health domain, information extraction in health, and ontology-based information extraction. Section 4 contains the general vision/ proposal of an ontology-based information extraction system to feed a search engine. The respective instantiation in two systems with different purposes and some illustrative results are presented in Section 5. Chapter ends with conclusions, provided in Section 6.
3.2 Background
Several approaches to IE have been followed over the years. One common approach is based on pattern matching and exploits basic patterns over a variety of structures: text strings, part-of-speech tags, semantic pairs, and dictionary entries (Pakhomov 2005). However, this type of approach does not generalize well, which limits its extension to new domains. The need for IE systems that can be easily adapted from one domain to another leads to the development of different approaches based on adaptive IE, starting with the Alembic Workbench (Aberdeen et al. 1995). The idea behind these approaches is to use various kinds of machine learning algorithms to allow IE systems to be easily targeted to new problems. The effort required to redesign a new system is replaced with that of generating batches of training data and applying learning algorithms.
A more recent approach is the ontology-based IE (OBIE), which aims at using ontology to guide the information extraction process (Hahn, Romacker & Schulz 2002). Since Berners-Lee et al. (1994) and Berners-Lee & Fischetti (1999) began to endorse ontologies as the backbone of the semantic web in the 1990s, a whole research field has evolved around the fundamental engineering aspects of ontologies, such as their generation, evaluation and management.
A relevant number of approaches need seed examples to train the IE systems. As such, several tools to annotate the semantic web were developed. Some earlier systems involved having humans annotate texts manually, using user-friendly interfaces (Handschuh, Staab & Studer 2003; Schroeter, Hunter & Kosovic 2003). Others featured algorithms to automate part of the annotation process. Those algorithms were based on manually constructed rules or extraction patterns, to be completed based on the previous annotations (Alfonseca & Manandhar 2002; Ciravegna et al. 2002).
As manual annotation can be a time consuming task, some approaches involved using ontology class and subclass names to generate seed examples for the learning process. Those names are used to learn contexts from the web and then those contexts are used to extract information (Kiryakov et al. 2004; Buitelaar et al. 2008). Other approaches added Hearst patterns to increase the amount of seed examples (McDowell & Cafarella 2008). For instance, considering the Bird class, useful patterns would be “birds such as X,” “birds including X,” “X and other birds,” and “X or other birds,” among others.
3.3 Related work
Relevant related work on search, particularly search applied to health information is the focus of this section. Here, relevant work related to ontology-based health information search is presented. First, recent trends in semantic search are presented; thereafter we discuss some of the trends related to health search and its specificities followed by a discussion of information extraction applied to health. The section ends with recent relevant work on OBIE.
3.3.1 Semantic search
In recent years, the interest in semantic search has increased. Even mainstream search engines such as Google or Bing are evolving to include semantics. Some systems do not assume that all, or most data, have a formal semantic annotation. One approach is expanding the user query by including synonyms and meronyms of the queried terms (Moldovan & Mihalcea 2000; Buscaldi, Rosso & Arnal 2005). Term expansion is made using the “OR” operation available in most search engines. A somewhat similar approach is followed by Kruse et al. (2005), which uses WordNet ontology and the “AND” operation of search engines to provide semantic clarification on concepts that have more than one meaning in WordNet.
Other approaches combine full text search and ontology search. ESTER (Bast et al. 2007) features an entity recognizer that assigns words or phrases to the entities of the ontology. Then, when searching for information, two basic operations are used: prefix search and join. This allows discrimination of different meanings of a concept but without logic inference. A different approach is adopted by Rocha, Schwabe & Aragao (2004). This approach involves using a regular full text search plus locating additional relevant information by using other document data such as document creator. This additional data is stored in a RDF graph, which is traversed in order to find similar concepts.
Systems that process only data with a formal semantic annotation use SPARQL queries to retrieve results (Guha & McCool 2003; Lei, Uren & Motta 2006; Esa, Taib & Thi 2010). The problems usually addressed in these cases are performance, in terms of reasoning speed, and how to rank the result set. A discussion on semantically enhanced search engines for web content discovery can be found in Kamath et al. (2013). Jindal, Bawa & Batra (2013) present a detailed review of ranking approaches for semantic search on web.
3.3.2 Health information search and exploration
In the health domain, web-available search engines are mainly targeted at retrieving information from related knowledge resources such as PubMed, the Medical Subject Headings thesaurus (MeSH) of the U.S. National Library of Medicine and the Unified Medical Language System (UMLS) of the U.S. National Library of Medicine.
CISMeF and HONselect are examples of such systems. The objective of CISMeF (Darmoni et al. 2000) is to assist health professionals and consumers in their search for electronic health information available on the Internet. CISMeF, initially only available in French, has recently improved in two ways, being currently: (1) a generic tool able to describe and index web resources and PubMed citations or Electronic Health Records; (2) multi-lingual by allowing queries in multiple terminologies and several languages. HONselect (Boyer et al. 2001) presents medical information arranged under MeSH, offering advanced multilingual features to facilitate comprehension of web pages in languages other than those of the user.
Another health-specific information search engine is WRAPIN (Gaudinat et al. 2006). WRAPIN combines search in medical Web pages with other “hidden” online documents that are not referenced by other search engines. WRAPIN analyses a page for the most important medical terms, performing frequency analysis on MeSH terms found on the page. It identifies keywords which are then used for weighted queries to its indexes and to translate into languages other than that of the initial query. WRAPIN also allows the most important medical concepts in the document to be highlighted.
Can & Baykal (2007) designed MedicoPort, a medical search engine designed for users with no medical expertise. It is enhanced with domain knowledge obtained from UMLS in order to increase search effectiveness. MedicoPort is semantically enhanced by transforming a keyword search into a conceptual search, both for web pages and user queries.
As an example of recent work, Mendonça et al. (2012) designed and developed a proof-of-concept system for a specific group of target users and a specific domain, namely, Neurological Diseases. The application allows users to search for neurologic diseases, and collects a set of relevant documents with the support of ontology navigation as an auxiliary tool to redefine a query and change previous results.
Another example of recent work is that of Dragusin et al. (2013) who introduced FindZebra, a specialized rare disease search engine powered by open-source search technology. FindZebra uses freely available online medical information, but also includes specialized functionalities such as exploiting medical ontological information and UMLS medical concepts to demonstrate different ways of displaying results to medical experts. The authors concluded that specialized search engines can improve diagnostic quality without compromising the ease of use of the current and widely popular web search engines.
3.3.3 Information extraction for health
In the clinical domain, IE was initially approached with complete systems, i.e., systems including all functions required to fully analyze free-text. Examples of these large-scale projects are:
- – The Linguistic String Project – Medical Language Processor of New York University
- – The Specialist system (McGray et al. 1987) developed at the United States National Library of Medicine as part of UMLS project. This system includes the Specialist Lexicon, the Semantic Network, and the UMLS Metathesaurus (USNLM 2008)
- – The Medical Language Extraction and Encoding system (MedLEE) system (Friedman et al. 1995) developed at the New York Presbyterian Hospital at Columbia University. MedLEE is mainly semantically driven; it is used to extract information from clinical narrative reports, to participate in an automated decision-support system, and to allow natural language queries.
Significant resources were required to develop and implement these complete medical language processing systems. Consequently, several authors experimented over time with simpler systems that were focused on specific IE tasks and on limited numbers of different types of information to extract. Some of the areas currently benefiting from IE methods are biomedical and clinical research, clinical text mining, automatic terminology management, decision support and bio-surveillance. These narrowly focused systems demonstrated such good performance that they now constitute the majority of systems used for IE. Relevant examples of this type of system are those from the International Classification of Diseases (ICD) (Aronson et al. 2007; Crammer et al. 2007).
3.3.4 Ontology-based information extraction – OBIE
Different approaches to OBIE have been proposed and developed over the years. The approaches differ in some dimensions as follows: (1) the identification and extraction of information can be performed using probabilistic methods or explicitly defined sets of rules; (2) the types of document from which information is extracted can be unstructured, plain text, or semi-structured and structured sources; (3) the ontology can be constructed from the document’s content or exist before the process started, and has the option of being updated automatically while processing documents; and (4) the kind of information extracted varies from extracting only ontological instances to extracting entire ontological classes and its associated properties.
Several IE groups focused on the development of extraction methods that use the content and predefined semantics of an ontology to perform the extraction task without human intervention and dependency on other knowledge resources (Embley et al. 1998; Maedche et al. 2002; Buitelaar & Siegel 2006; Yildiz 2007).
Considering IE for generic domains, a frequent approach is to use Wikipedia to build their knowledge base. Relevant examples of such systems were developed by Bizer et al. (2009), Suchanek, Ifrim & Weikum (2007), and Wu, Hoffmann & Weld (2008). Wikipedia structure is used to infer the semantics, and the knowledge base is populated by extracting information from pages of text and infoboxes. Other approaches, that do not take advantage of Wikipedia structure, acquire information from generic web pages. The knowledge base structure is often inferred from pages of content and the knowledge base is populated using the same sources. Two good examples are Etzioni et al. (2004) and Yates et al. (2007).
Most approaches, whether using Wikipedia or not, use shallow linguistic analysis to detect the information to extract. Shallow analysis involves detecting text patterns and, at most, using part-of-speech information: e.g., which words are nouns, verbs, adverbs, or adjectives. The use of shallow linguistic information, however, makes it difficult to acquire information from complex sentences. A comprehensive survey of current approaches to OBIE can be found in Wimalasuriya & Dou (2010).
3.4 A general architecture for health search: handling both private and public content
Health-related information can have quite different access restrictions. Personal health-related information is confidential and has restricted access even inside health organizations. On the other hand, other health-related information, such as general information on drugs and disease characterization, is available to the general public. Not every case falls neatly into either personal health information that is confidential or more broadly into the category of general health info accessible to the public writ large. It is thus important that both well-defined cases which are either personal health-related or general-health related, and those that are not as extreme but share features of both categories, be addressed in a similar way. In this section we present a unified view of these cases (Fig. 3.1).
The main differences between the two extreme cases of personal health info versus general health pivots on (1) who are the target users of the information, i.e., if the search is made available to a general audience or only to authorized users (power users in the figure); and (2) if such a search is only possible inside the intranet of the organization holding the original source of information. In our view, these differences do not need different architectures to be handled effectively. In both cases, processing of the public and private documents can be performed by a similar pipeline, combining IE and semantic integration, and making use of ontologies. The Search and Inference Engine can also be the same. The only requirement is that the interfaces can handle access restrictions, preventing both unauthorized user access and access from the web when the information is for an organization’s restricted internal use.
The proposed architecture is composed of a set of modules in which the IE component consists of a basic set of processing elements similar to the basic set illustrated in Fig. 3.2 and described by Hobbs (2002). The semantic integration module is responsible for merging the information extracted in the previous modules and reasoning. The following sections describe in detail the architecture of the two different systems used in this work.
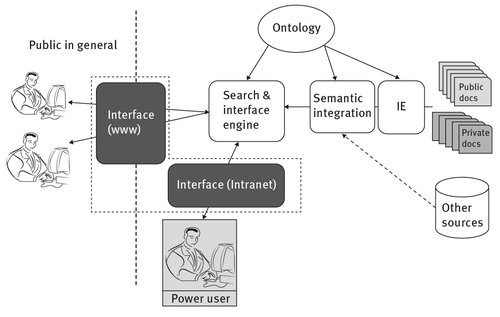
Fig. 3.1: Unified view of semantic search for health, handling both access by the general public and restricted access by authorized users inside an organization.
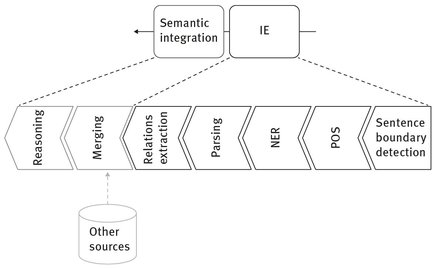
Fig. 3.2: The basic set of modules to be included in the processing pipeline to extract information from natural language sources and feeding the search engine.
3.5 Two semantic search systems for health
In this section we present in detail two different systems providing semantic search supported by information extraction for both private and public content. The first system targets the search inside a health institution such as a hospital, or more generally what is considered an Intranet search, which is a search within an organization’s own internal website or group of websites. The second system in contrast targets the search outside the confines of the organization so as to enable the general public to semantically search and explore health-related information made available on the World Wide Web in Portuguese. Both of these semantic search systems were designed for Portuguese, but can be readily extended to other languages.
3.5.1 MedInX
MedInX (Ferreira, Teixeira & Cunha 2012) is a medical information eXtraction system tailored to process textual clinical discharge records in order to perform automatic and accurate mapping of free text reports onto a structured representation. MedInX is designed to be used by health professionals, and by hospital administrators and managers, as it also allows its users to search the contents of such automatically populated ontologies. MedInX uses IE technology to structure the information present in discharge reports originated by the electronic health record (EHR) system used in the region of Aveiro, Portugal in the Telematic Healthcare Network RTS® (Cunha et al. 2006). The way it works is by automatically instantiating a knowledge representation model from the free-text patient discharge letters (PDL) issued by the hospital.
During a patient’s hospitalization, a large amount of data is produced in textual form as in the case of patient discharge letters. The purpose of these documents is to transfer summarized information from the hospital setting to other places, normally to the general practitioner, in order to assure continuity of patient care. MedInX addresses this type of narrative since they cover the whole inpatient period and summarize the major occurrences during that period.
The first step in development of MedInX was the creation of a corpus of authentic health records to be used in the development and evaluation of the system. This corpus was gathered through a list of hospital episodes for which a code had been assigned relative to the diagnosis of a cerebrovascular disease. If more than one cerebrovascular disease were found in the patient, additional codes pertaining to those conditions were entered in the patient’s record. The corpus consists, thus, of 915 discharge letters written in Portuguese, corresponding to patients admitted with at least one of the several cerebrovascular disease-related codes. Table 3.1 gives statistics pertaining to the amount of documents, sentences, and tokens included in the development and test-evaluation set of MedInX.
Tab. 3.1: Number of documents, tokens and sentences in the MedInX corpus and its subsets.
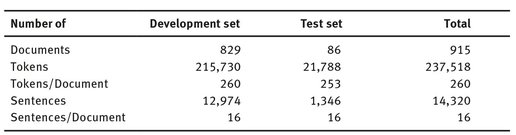
Figure 3.3 presents a style-preserving illustration of a PDL, showing how it is possible to analyze the general content and structure of the documents. To begin with, the discharge documents have several interesting contextual features. In general, it is evident that the narratives are written from one professional to another in order to support information transfer, remind them about important medical facts, and supplement with crucial numerical data such as blood pressure and lab test results. The texts are normally intelligible and the meaning becomes evident from the context even in the presence of numerous linguistic and grammatical mistakes, word abbreviations, acronyms, signs, and other communicative features.
MedInX is a system designed for the clinical domain, which contains components for the extraction of hypertension-specific characteristics from unstructured PDLs. Its components are based on NLP principles; as such, they contain several mechanisms to read, process, and utilize external resources, such as terminologies and ontologies. These external resources represent an important part of the system by providing structured representations of the domain, clinical facts, and events that are present in the texts. The MedInX ontologies allow the assignment of domain-specific meanings to terms and use these meanings in their operations.
3.5.1.1 MedInX ontologies
In MedInX four new ontologies were created. The first two consist of two formalizations of the international classification systems that are supported by the World Health Organization (WHO): the International Classification of Diseases (ICD); and the International classification of functioning, disability and health (ICF). A drugs ontology and a conceptualization of the structure and content of the discharge reports comprise the last two of the MedInX ontologies.

Fig. 3.3: A style-preserving illustration of a patient discharge letter. The original document written in portuguese is presented on the left of the figure and, on the right, its English translation.
This last ontology, in particular, was designed as an extensible knowledge model and used for storing and structuring the PDLs’ entities and their relations, including temporal and modifying information. For instance, the MedInX ontology describes the fact that a Sign or Symptom is a Condition, a prescribed Medication is a Therapeutic, a Procedure can be targeted to an Anatomical Site, and so forth. To identify the concepts in the ontology a middle-out strategy was used, i.e., first the core basic terms were identified in text and then specified and generalized as required.
3.5.1.2 MedInX system
MedInX components run within the unstructured information management architecture (UIMA) framework (Ferrucci & Lally 2004). Figure 3.4 illustrates MedInX architecture, identifying the following components:
- Document Reader: a component which converts PDL files into plain text and extracts implicit meaning from the structure of the document by converting the embedded tags of the input document into annotations;
- General natural language processing: components for sentence discovery, tokenization and part-of-speech tagging;
- REMMIX: the Named Entity Recognition component of MedInX which concentrates on the later stages of IE, i.e., takes the linguistic objects as input and finds domain-dependent classifications and patterns among them. REMMIX is made up of three other components:
- Context Dependent annotator: an annotator that creates annotations from one or more tokens, using regular expressions and surrounding tokens as clues;
- Concept finding: a component which extracts concepts based on specified terminologies and ontologies, and determines negation, lateralization and modifiers;
- Relation extraction: a component which extracts relations between concepts using contextual information;
- Consumers: responsible for ending the process and creating the desired output. The three main consumers of MedInX are:
- XML consumer: which produces an XML file with the annotations of the previous annotators.
- Ontology population: which populates the MedInX ontology. This component produces an OWL file with the information extracted.
- Template filling: which outputs the knowledge extracted from the narratives to a template containing information about the patient and their health related state, with the correspondent ICF codes.
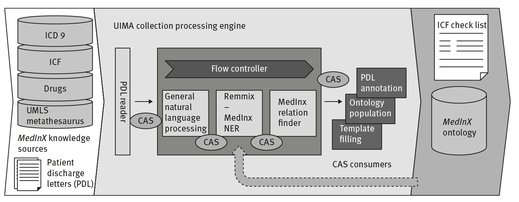
Fig. 3.4: MedInX architecture.
3.5.1.3 Representative results
Figure 3.5 presents MedInX evaluation interface. The extracted entities are identified by the filled boxes while the arrows represent the relations between these.
MedInX was first evaluated in 2011, in the task of extracting information from PDLs. Seven judges, belonging to different specialized areas ranging from medicine to computer science to linguistics, participated in the assessment by using the web-based evaluation interface (Fig. 3.5). To wit, the jury was made up of two computer scientists, a linguist, a radiologist, two psychologists and a physician. The 86 PDLs of the evaluation set initially selected from the MedInX corpus were automatically annotated by MedInX and made available for evaluation for four months. During this four-month period a total of 30 different reports were reviewed.
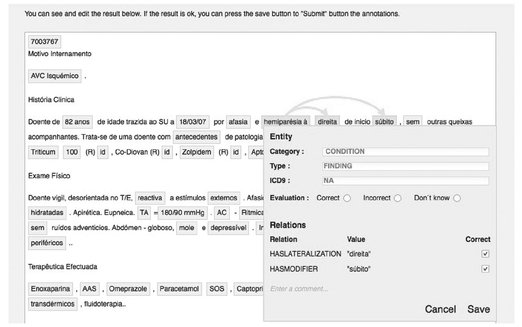
Fig. 3.5: MedInX evaluation interface.
The results obtained in the task of semantic classification are presented in Tab. 3.2 in terms of precision, recall, and F-measure. These values indicate a good performance of MedInx all the way around. For example, given that MedInX is tailored to the medical domain and intended to process clinical text, its proficiency with correctly extracting the entities and relations described in text makes it very well suited to the task. That is, only a precise system is capable of producing a correct, consistent, and concise ontology. Nonetheless, we were also concerned with the completeness of such an ontology, i.e., with the recall of the system. All in all, the results obtained indicate that MedInX performs with both high precision and recall, each showing an evaluation at approximately 95%. This is supported by an F-Measure, the Harmonic mean of recall and precision, whose evaluation is likewise at approximately 95%.
The clinical data included in the PDLs is a rich source of information, not only about the patient’s medical condition, but also about the procedures and treatments performed in the hospital. Searching the content of the PDLs and ensuring the completeness of these documents is a process that still needs to be performed manually by expert physicians in health institutions. In order to support this process, we developed the MedInX clinical audit system. The main objective of the audit system is to help not only physicians, but hospital administrators and managers as well, to access the contents of the PDLs.
The clinical audit system uses the automatically populated MedInX ontology, which contains the structured information automatically extracted from the PDLs, and performs an automatic analysis of the content and completeness of the documents. The MedInX audit system uses the RDF query language SPARQL to query the ontology and retrieve relevant information from this resource. Several levels of information can be retrieved from this resource. An example of a developed rule, describing a complex scenario is given in Fig. 3.6. With this rule, we can find the PDLs that refer to less than a number of clinical Conditions and over a certain number of Chemical Procedures, namely, Medications and Active Substances. Both numbers used by the rule are user defined. The example of the figure uses the value 17 as the defined threshold after analysis of the most common values for these entities in the PDLs. The result of this rule allows identification of the outlier reports and suggests the need for content verification.
Tab. 3.2: Results of MedInX in the task of semantic classification.

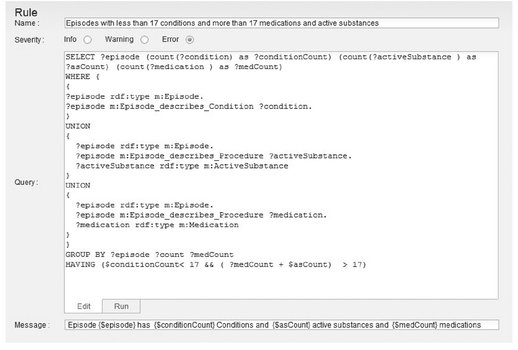
Fig. 3.6: MedInX audit system rule editor.
3.5.2 SPHInX – Semantic search of public health information in portuguese
The proof of concept system described next aims to perform semantic search on health information publicly available on the web in Portuguese.
3.5.2.1 System architecture
The SPHInX implementation is organized in four modules, respectively:
- – Natural Language Processing: includes document content processing technologies to retrieve structured information from natural language texts. It is named NLP because it is based on technologies from the NLP area. In this part of the prototype, text is extracted from documents and enriched with the inclusion of POS tags, identification of named entities, and the construction of syntactic structures.
- – Domain Representation: has tools for defining data semantics and associates it with samples of the NLP module output. System semantics is defined via ontology and, according to the ontology defined, it is necessary to provide examples of ontological classes and relations in sample documents. The examples are used to train semantic extraction models.
- – Semantic Extraction and Integration: trains and applies semantic extraction models to all texts in order to obtain meaningful semantic information. It complements the extracted information with external structured sources, e.g., geocodes and stores everything in a knowledge base conforming to the defined ontology.
- – Search: information in the knowledge base can be searched and explored using natural language queries or via SPARQL.
3.5.2.2 Natural language processing
SPHInX was developed to process generic unstructured documents written in Portuguese, not only PDLs. Thus the NLP part was developed to handle the Portuguese language. The processing is organized in four sequential steps: (1) end of sentence detection; (2) Part-of-Speech (POS) tagging; (3) Named Entity Recognition (NER); and (4) syntactic parsing.
Text is extracted from documents, and then sentences are separated using the sentence boundary detector Punkt (Kiss & Strunk 2006). The sentence boundary detector step is highly relevant because all natural language processing is done in a per sentence fashion. This means that sentences define the processing context in the next NLP steps, which include algorithms able to use all the content of a sentence without using any content of the previous or following sentences.
After the split, sentences are enriched with POS tags assigned by TreeTagger (Schmid 1994): noun, verb, adjective, etc. TreeTagger was trained with a European Portuguese lexicon in order to be integrated in the system. Its outputs contain the word form followed by the assigned POS tag and the word lemma.
Named entities are discovered and classified by REMBRANDT (Cardoso 2012). Words belonging to a named entity are grouped using underscores. For instance, the names of the person John Stewart Smith become the single token John_ Stewart_Smith. Then, sentences are analyzed to determine their grammatical structure. This is done by MaltParser (Hall et al. 2007) and the result is a planar graph encoding the dependency relations among the words of each sentence.
3.5.2.3 Semantic extraction models
SPHInX creates one semantic extraction model for each ontology class and ontology relation. A model is a set of syntactic structure examples and counter examples that were found to encode the meaning represented by the model. It also contains a statistical classifier that measures the similarity between a given structure and the model’s internal examples. The model is said to have positively evaluated a sentence fragment if the similarity is higher than a given threshold.
The algorithm for creating semantic extraction models was inspired in two studies. The first is about extracting instances of binary relations using deep syntactic analysis. Suchanek, Ifrim & Weikum (2006) extracted one-to-one and many-to-one relations such as place and date of birth. They used custom-built decision functions to detect facts for each relation, and a set of statistical classifiers to decide if new patterns are similar to the learned facts. In our proof-of-concept prototype, this work was extended to include the extraction of one-to-many and many-to-many relations. The proof-of-concept prototype also implements a general purpose decision function based on the annotated examples instead of a custom-built function for each relation.
The second work is about improving entity and relation extraction when the process is learned from a small number of labeled examples, using linguistic information and ontological properties (Carlson et al. 2009). Improvements are made using class and relation hierarchy, information about disjunctions, and confidence scores of facts. This information is used to bootstrap more examples thereby generating more data to train statistical classifiers. For instance, when the system is confident about a fact, such as when it was annotated by a person, this fact is used as an instance of the annotated class and/or relation. This fact can also be used as a counter-example of all classes/relations disjoint with the annotated class/relation, and as an instance of super-class/super-relation. Moreover, facts discovered by the system with a high confidence score can be promoted to examples and included in a new round of training. In the proof-of-concept prototype, this creation of more examples is not active by default as it can lead to data over-fitting and should therefore be used carefully.
For the first version of SPHInX, the ontology about neurological diseases used in Mendonça et al. (2012) was adopted. The semantic extraction models were trained with a set of six manually annotated documents, of around fifty pages each, by a person familiar with the ontology but not related to the prototype development. The annotations were related to neurological diseases and respective symptoms, risk factors, treatments and related drugs.
3.5.2.4 Semantic extraction and integration
All sentence graphs are evaluated by the classifiers of all semantic models, and are collected in the case of forming a triple. A sentence fragment forms a triple if it is positively evaluated by two class models, one for subject and the other for object, along with one relation model binding the subject and object (Rodrigues, Dias & Teixeira 2011). Missing information according to the ontology is searched in external structured information sources. For instance, unknown locations of entities with a fixed place (such as streets, organizations’ headquarters, and some events) are queried using Google Maps API.
All collected triples are tentatively added to the knowledge base and their coherence is verified by a semantic reasoner. In SPHInX, reasoning is performed by an open-source reasoner for OWL-DL named Pellet (Sirin & Parsia 2004). All triples not coherent with the rest of the knowledge base are discarded, and a warning is issued. The remaining triples become part of the knowledge base.
3.5.2.5 Search and exploration
The search and exploration part of the system will be explained based on an illustrative example of use. The data for this example was obtained by having the system process fourteen previously unseen documents using the semantic extraction models trained earlier, plus the data already on the ontology at the time. Then, the same person that annotated the training documents was asked to suggest a few possible questions in Portuguese. Those questions were submitted to the system and one of them was selected for the example.
The interface, based on NLP-Reduce (Kaufmann, Bernstein & Fischer 2007), accepts natural language questions and generates SPARQL queries that are passed to a SPARQL engine. The system allows the user to enter a sentence, such as “memory loss is a symptom of what diseases?”
First, the question is transformed by removing all stop words and punctuation marks. The remaining words are stemmed and passed to a query generator that will use them to produce a SPARQL query in four steps:
- Search for triples that contain one or more words of the query in the object property label. Triples are ranked according to the amount of words included in the label.
- Search for properties that can be joined with the triples found in step 1. Thus, properties are searched using domain and range information of triples from step 1 along with the remaining query words. In the case of query words producing triples based on alternative object properties, the triples favored are those with the highest score from step 1. The triple set of this step is combined with the set of step 1, according to the ontology rules.
- Search for data type property values that match the query words not matched in steps 1 and 2. Triples found are once again ranked considering the amount of words included in the property values. All triples found respecting the domain and range restrictions of the set created in step 2 are added to it.
- When there are no more query words left, the SPARQL query is generated to join the retrieved triples that achieved the highest scores in steps 1 to 3. Semantically equivalent duplicates are removed and the query is ready to be passed to a SPARQL endpoint.
So a question like “a perda de memória é um sintoma de que doenças?” which is roughly the Portuguese equivalent to “memory loss is a symptom of what diseases?” originates the SPARQL code presented in Fig. 3.7.
The output of the query is a table containing the variables of the SPARQL query. Depending on the type of information asked, the data on the table can be plain text as in case of data type property, presented in Tab. 3.3, or links to other ontological entities as in the case of object properties. In the case of the latter, it is then possible to navigate through the ontology by following those links.
Another way to output results is by presenting the graph of ontological concepts involved in the query. Figure 3.8 depicts the graph of the ontological elements used to compute the answer to the example query. As can be seen, there are more concepts involved than the ones included in the output table. The concepts involved in queries and not presented in the output table are typically the ones used to compute logical inferences.
Presenting the results in a graphic format allows users to navigate the information stored in the knowledge base while keeping a good overview of the ontology and how different concepts relate to each other.

Fig. 3.7: SPARQL code generated by the natural language interface.
Tab. 3.3: Result set for the query “a perda de memória é um sintoma de que doenças?” (“memory loss is a symptom of what diseases?”).
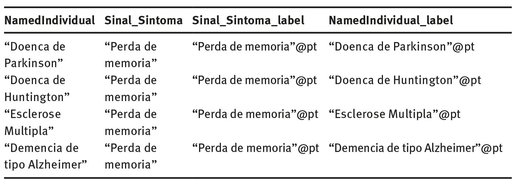
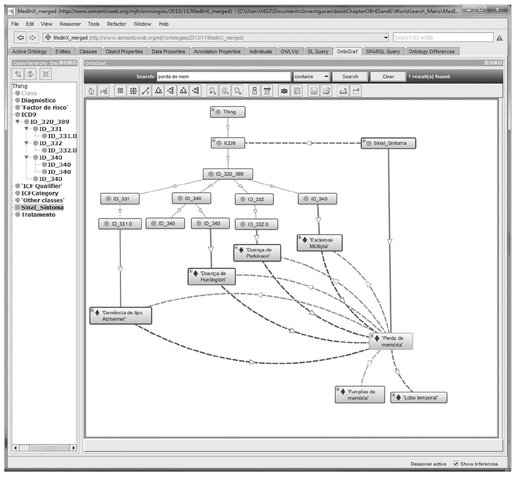
Fig. 3.8: Graph of the ontology concepts involved in the query example.
3.6 Conclusion
Taking into consideration the increasing need for semantic search of health information available originally in natural language, in this chapter a general architecture predicated on ontology-based information extraction to feed a search engine is proposed and instantiated in two systems. The first system, MedInx, allows semantic search of the information regarding a hospital’s discharge letters, and can be generalized to the vast information in natural language stored in internal web-based hospital information systems. The second system, SPHInX, currently at an early stage of development, is capable of extracting information from public documents in Portuguese. For both systems, we present information on its architecture and components, and show via demonstration how these systems work.
We envision future developments of both systems that would address a much broader area, as both systems that we presented here only address a limited medical domain. Another equally important goal pivots on the improvement of the interaction of the user with these systems, making search and exploration a natural experience for professionals and laity alike.
Acknowledgments
This work was partially supported by World Search, a QREN project (QREN 11495) co-funded by COMPETE and FEDER, and the Portuguese Foundation for Science and Technology PhD grant SFRH/BD/27301/2006 to Liliana Ferreira. The authors also acknowledge the support from IEETA Research Unit, FCOMP-01-0124-FEDER-022682 (FCT-Pest C/EEI/UI0127/2011).
References
Aberdeen, J., Burger, J., Day, D., Hirschman, L., Robinson, P. & Vilain, M. (1995) MITRE: description of the Alembic system used for MUC-6. Proceedings of the 6th conference on Message understanding (pp. 141–155). New York, NY: Association for Computational Linguistics.
Abraham, J. & Reddy, M. (2007) Quality of healthcare websites: A comparison of a general-purpose vs. domain-specific search engine. AMIA Annual Symposium Proceedings, 858.
Aronson, A., Bodenreider, O., Demmer-Fushman, D., Fung, K., Lee, V. & Mork, J. (2007) From indexing the biomedical literature to coding clinical text. BioNLP ’07: Proceedings of the Workshop on BioNLP 2007, (pp. 105–112).
Bast, H., Chitea, A., Suchanek, F. & Weber, I. (2007) ESTER: Efficient Search on Text, Entities, and Relations. Proceedings of the 30th ACM SIGIR, (pp. 679–686).
Berners-Lee, T., Cailliau, R., Luotonen, A., Nielsen, H. F. & Secret, A. (1994) ‘The World-Wide Web’, Commun ACM, 37:76–82.
Berners-Lee, T. & Fischetti, M. (1999) Weaving the Web: The Original Design and Ultimate Destiny of the World Wide Web by Its Inventor. San Francisco: Harper Collins.
Bizer, C., Lehmann, J., Kobilarov, G., Auer, S., Becker, C., Cyganiak, R. & Hellmann, S. (2009) DBpedia – A crystallization point for the Web of Data. Web Semantics: Science, Services and Agents on the World Wide Web – The Web of Data, 7, 154–165.
Boyer, C., Baujard, V., Griesser, V. & Scherrer, J. R. (2001) ‘HONselect: a multilingual and intelligent search tool integrating heterogeneous web resources’, Int J Med Inform, 64:253–258.
Buitelaar, P. & Siegel, M. (2006) Ontology-based Information Extraction with SOBA. Proceedings of the International Conference on Language Resources and Evaluation (LREC), (pp. 2321–2324).
Buscaldi, D., Rosso, P. & Arnal, E. S. (2005) A wordnet-based query expansion method for geographical information retrieval. Working Notes for the CLEF Workshop.
Can, A. B. & Baykal, N. (2007) ‘MedicoPort: a medical search engine for all’, Comput Meth Prog Biomed, 86:73–86.
Cardoso, N. (2012) ‘Rembrandt – a named-entity recognition framework’. LREC, (pp. 1240–1243).
Carlson, A., Betteridge, J., Hruschka, E. R. & Mitchell, T. M. (2009) Coupling Semi-Supervised Learning of Categories and Relations. SemiSupLearn ’09: Proceedings of the NAACL HLT 2009 Workshop on Semi-Supervised Learning for Natural Language Processing (pp. 1–9). Association for Computational Linguistics.
Cowie, J. & Lehnert, W. (1996) ‘Information extraction’, Communications of the ACM, 39:80–91.
Crammer, K., Dredze, M., Ganchev, K., Talukdar, P. P. & Carroll, S. (2007) Automatic code assignment to medical text. BioNLP ’07: Proceedings of the Workshop on BioNLP 2007 (pp. 129–136). Association for Computational Linguistics.
Cunha, J. P., Cruz, I., Oliveira, I., Pereira, A. S., Costa, C. T., Oliveira, A. M. & Pereira, A. (2006) The RTS project: Promoting secure and effective clinical. eHealth 2006 High Level Conference, (pp. 1–10).
Darmoni, S. J., Leroy, J. P., Baudic, F., Douyère, M. A. & Thirion, B. (2000) ‘CISMeF: a structured health resource guide’, Met Inform Med, 30–35.
Dragusin, R., Petcu, P., Lioma, C., Larsen, B., Jorgensen, H. L., Cox, I. J., Hansen, L. K., Ingwersen, P. & Winther, O. (2013) ‘FindZebra: a search engine for rare diseases’, Int J Med Inform, 82:528–538.
Embley, D. W., Campbell, D. M., Smith, R. D. & Liddle, S. W. (1998) Ontology-based extraction and structuring of information from data-rich unstructured documents. Proceedings of the seventh international conference on Information and knowledge management (pp. 52–59). ACM.
Esa, A. M., Taib, S. M. & Thi, H. N. (2010) Prototype of semantic search engine using ontology. Open Systems (ICOS), 2010 IEEE Conference on (pp. 109–114). IEEE.
Etzioni, O., Cafarella, M., Downey, D., Kok, S., Popescu, A.-M., Shaked, T., Soderland, S., Weld, D. S. & Yates, A. (2004) Web-Scale Information Extraction in KnowItAll (Preliminary Results). WWW ’04 – Proceedings of the 13th International World Wide Web Conference (pp. 100–110). New York, NY, USA: Association for Computational Linguistics.
Ferreira, L., Teixeira, A. & Cunha, J. P. (2012) Medical Information Extraction – Information Extraction from Portuguese Hospital Discharge Letters. Lambert Academic Publishing.
Ferrucci, D. & Lally, A. (2004) ‘UIMA an architectural approach to un-structured information’, Nat Lang Eng, 10:327–348.
Friedman, C., Johnson, S. B., Forman, B. & Starren, J. (1995) ‘Architectural requirements for a multipurpose natural language processor in the clinical environment’. Proc Annu Symp Comput Appl Med Care, (pp. 347–351).
Gaizauskas, R. & Wilks, Y. (1998) ‘Information extraction: beyond document retrieval’, J Doc, 54:70–105.
Gaudinat, A., Ruch, P., Joubert, M., Uziel, P., Strauss, A., Thonnet, M., Baud, R., Spahni, S., Weber, P., Bonal, J., Boyer, C., Fieschi, M. & Geissbuhler, A. (2006) ‘Health search engine with e-document analysis for reliable search results’, Int J Med Inform, 75:73–85.
Gruber, T. R. (1993) ‘A translation approach to portable ontology specifications’, Knowledge Acquisition, 5:199–220.
Guarino, N. (1998) Formal Onthology in Information Systems. FIOS’98 – Proceedings of the First International Conference on Formal Ontology in Information Systems (pp. 3–15). IOS Press.
Guha, R. & McCool, R. (2003) ‘TAP: a Semantic Web platform’, Computer Networks, 557–577.
Guha, R., McCool, R. & Miller, E. (2003) Semantic search. Proceedings of the 12th international conference on World Wide Web (pp. 700–709). ACM.
Hahn, U., Romacker, M. & Schulz, S. (2002) ‘Creating Knowledge Repositories From Biomedical Reports: The MEDSYNDIKATE Text Mining System’. Pac Symp Biocomput, (pp. 338–349).
Hall, J., Nilsson, J., Nivre, J., Eryigit, G., Megyesi, B., Nilsson, M. & Saers, M. (2007) Single Malt or Blended? A Study in Multilingual Parser Optimization. (pp. 933–939). Association for Computational Linguistics.
Hobbs, J. R. (2002) ‘Information extraction from biomedical text’, J Biomed Inform, 35:260–264.
Jindal, V., Bawa, S. & Batra, S. (2014) ‘A review of ranking approaches for semantic search on Web’. Inf Process Manage, 50(2): 416–425.
Kamath, S. S., Piraviperumal, D., Meena, G., Karkidholi, S. & Kumar, K. (2013) A semantic search engine for answering domain specific user queries. Communications and Signal Processing (ICCSP), 2013 International Conference on (pp. 1097–1101). IEEE.
Kaufmann, E., Bernstein, A. & Fischer, L. (2007) NLP-Reduce: A “naive” but Domain-independent Natural Language Interface for Querying Ontologies. ESCW’07 – Proceedings of the 6th International Semantic Web Conference.
Kiss, T. & Strunk, J. (2006) ‘Unsupervised multilingual sentence boundary detection’, Compu Linguist, 32:485–525.
Kruse, P. M., Naujoks, A., Rosner, D. & Kunze, M. (2005) Clever search: A wordnet based wrapper for internet search engines. arXiv preprint cs/0501086.
Lee, L. (2004) “I’m sorry Dave, I’m afraid I can’t do that”: Linguistics, Statistics, and Natural Language Processing circa 2001. In C. O. Board, Computer Science: Reflections on the Field, Reflections from the Field (pp. 111–118). Washington DC: The National Academies Press.
Lei, Y., Uren, V. & Motta, E. (2006) Semsearch: A search engine for the semantic web. Proceedings of the 15th International Conference on Managing Knowledge in a World of Networks Berlin, Heidelberg (pp. 238–245). Berlin, Heidelberg: Springer-Verlag.
Maedche, A., Maedche, E., Neumann, G. & Staab, S. (2003) Bootstrapping an Ontology-based Information Extraction System, pp. 345–359. Heidelberg, Germany: Physica-Verlag GmbH.
Màrquez, L., Carreras, X., Litkowski, K. C. & Stevenson, S. (2008) ‘Semantic role labeling: an introduction to the special issue’, Comput Linguist, 34:145–159.
McGray, A. T., Sponsler, J. L., Brylawski, B. & Browne, A. (1987) ‘The role of lexical knowledge in biomedical text understanding’. SCAMC, (pp. 103–107).
McNaught, J. & Black, W. (2006) Information extraction. In Ananiadou, S. & McNaught, J. Text Mining for Biology and Biomedicine (pp. 143–178). Norwood: Artech House.
Mendonça, R., Rosa, A. F., Oliveira, J. L. & Teixeira, A. J. (2012) Towards Ontology Based Health Information Search in Portuguese – A case study in Neurologic Diseases. CISTI’2012 – 7th Iberian Conference on Information Systems and Technologies.
Miller, G. A., Beckwith, R., Fellbaum, C., Gross, D. & Miller, K. J. (1990) ‘Introduction to wordnet: An on-line lexical database’, Int J Lexico, 235–244.
Moldovan, D. I. & Mihalcea, R. (2000) ‘Using wordnet and lexical operators to improve internet searches’, Internet Comput, IEEE, 4:34–43.
Pakhomov, S. A. (2005) High throughput modularized NLP system for clinical text. Proceedings of the ACL 2005 on Interactive poster and demonstration sessions (pp. 25–28). New York, NY: Association for Computational Linguistics.
Rocha, C., Schwabe, D. & Aragao, M. P. (2004) A hybrid approach for searching in the semantic web. Proceedings of the 13th international conference on World Wide Web (pp. 374–383). ACM.
Rodrigues, M., Dias, G. P. & Teixeira, A. (2011) Ontology Driven Knowledge Extraction System with Application in e-Government. Proc. of the 15th APIA Conference, (pp. 760–774).
Schmid, H. (1994) Probabilistic Part-of-Speech Tagging Using Decision Trees. Proceedings of the International Conference on New Methods in Language Processing.
Sirin, E. & Parsia, B. (2004) Pellet: An OWL DL Reasoner, In Haarslev, V. and Möller, R. (eds), International Workshop on Description Logics (DL’04), pp. 212–213. British Columbia, Canada: Whistler.
Suchanek, F. M., Ifrim, G. & Weikum, G. (2006) LEILA: Learning to Extract Information by Linguistic Analysis. Proceedings of the 2nd Workshop on Ontology Learning and Population: Bridging the Gap between Text and Knowledge (pp. 18–25). New York, NY: Association for Computational Linguistics. Sydney, Australia
Suchanek, F. M., Kasneci, G. & Weikum, G. (2007) YAGO: A Core of Semantic Knowledge Unifying WordNet and Wikipedia. WWW ’07 – Proceedings of the 16th International World Wide Web Conference (pp. 697–706). New York, NY: Association for Computational Linguistics.
USNLM. (2008) UMLS Knowledge Sources. United Stated National Library of Medicine.
Wang, L., Wang, J., Wang, M., Li, Y., Liang, Y. & Xu, D. (2012) ‘Using internet search engines to obtain medical information: a comparative study’. J Med Internet Res, 14.
Wang, C., Xiong, M., Zhou, Q. & Yu, Y. (2007) PANTO: A Portable Natural Language Interface to Ontologies. ESWC2007 – Proceedings of the 4th European Semantic Web Conference (pp. 473–487). Berlin/Heidelberg: Springer.
Wimalasuriya, D. C. & Dou, D. (2010) ‘Ontology-based information extraction: An introduction and a survey of current approaches’, J Inform Sci, 36:306–323.
World Health Organization. (n.d.) International Classification of Diseases. Accessed on 01/27/2014, http://www.who.int/classifications/icd/en/
Wu, F., Hoffmann, R. & Weld, D. S. (2008) Information Extraction from Wikipedia: Moving Down the Long Tail. KDD ’08 – Proceedings of the 14th ACM SIGKDD International Conference on Knowledge Discovery and Data Mining (pp. 731–739). New York, NY: Association for Computational Linguistics.
Yates, A., Banko, M., Broadhead, M., Cafarella, M. J., Etzioni, O. & Soderland, S. (2007) TextRunner: Open Information Extraction on the Web. NAACL-HLT (Demonstrations) – Proceedings of Human Language Technologies: The Annual Conference of the North American Chapter of the Association for Computational Linguistics (pp. 25–26). Morristown, NJ: Association for Computational Linguistics.
Yildiz, B. (2007) Ontology-driven Information Extraction. PhD Thesis. Vienna University of Technology.
