13
MSDTrA: A Boosting Based-Transfer Learning Approach for Class Imbalanced Skin Lesion Dataset for Melanoma Detection
Lokesh Singh*, Rekh Ram Janghel and Satya Prakash Sahu
Department of Information Technology, National Institute of Technology, Raipur, India
Abstract
Pigmented skin lesion datasets comprise a higher percentage of benign lesion than the malignant lesions which lead to the class skewness issue in the dataset. Classifiers trained for analyzing the automated dermatoscopic pigmented lesions often suffer from data scarcity. Transfer learning permits to leverage the knowledge from the source domain to train a classifier towards the target domain when the data is rare. Importing knowledge from multiple or several sources towards increasing the chance of searching a source closer to a target may alleviate the negative transfer. A framework is proposed in this work to transfer knowledge from multiple different sources utilizing AdaBoost, TrAdaBoost and MultiSource Dynamic TrAdaBoost (MSDTrA), for melanoma detection. The effectiveness of the proposed framework is evaluated on four benchmark skin lesion datasets namely, PH2, ISIC16, ISIC17, and HAM1000 which demonstrate promising performance by alleviating negative-transfer by increasing multiple different sources.
Keywords: Dermoscopic, classification, melanoma, class imbalance, boosting, sampling
13.1 Introduction
One of the most lethal types of skin cancer called malignant melanoma is responsible for the wide majority of skin cancer deaths. Cancer is an ailment of the rampant growth of abnormal cells, which are the rudimentary blocks of the body [1, 2]. The body persistently develops novice cells for rapid growth, exchange tattered tissues and repair injuries. Moreover, cells multiply themselves and die in an orderly fashion. But sometimes the process of cells gets disturbed, neither they grow nor die in an orderly fashion. Due to which the body’s lymph fluid becomes abnormal and as a result, tumor arises. A tumor might be benign or malignant. A benign tumor is not cancerous while malignant is, as it spread themselves in other parts of the body via lymph fluid [3]. Melanoma occurs over those parts of the body which are exposed to the sun, but sometimes very rarely it appears anywhere in the body which are not at all exposed to the sun like inside the mouth, eyes, nervous system, etc. [4, 5]. From the latest reports, the melanoma deaths have cross over 20,000 early in Europe. Nevertheless, if we diagnosed early to melanoma, it becomes the most treatable type of cancer in the world [6]. In skin lesion detection, benign lesions are found in majority while malignant lesions are rare in the dataset [7], which makes the melanoma detection challenging with skewed class distribution. In account of size of data and skewness in class distribution datasets are categorized in four sections as shown in Figure 13.1. The skin lesion datasets used in this experimentation are small in size and imbalanced as well thus, falls under the category of ‘absolute rarity’.
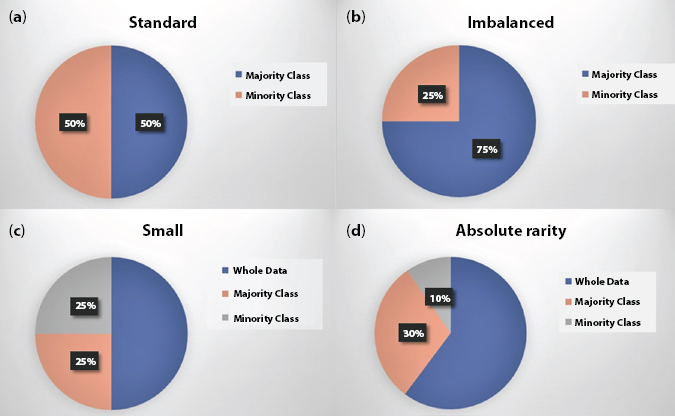
Figure 13.1 Different types of datasets: (a) standard (balanced), (b) unbalanced, (c) small-size, and (d) absolute-rare.
- A. Standard Dataset: This includes the adequate number of data within each class in a balanced manner i.e., each class contains an almost equal amount of data.
- B. Imbalanced Dataset: This includes an adequate number of data of one class called majority class and other class with fewer data in it generally known as a minority class.
- C. Small Dataset: This type of dataset includes the exiguous data in both the classes but is balanced.
- D. Rare Dataset: This type of dataset contains the exiguous data in an unbalanced manner i.e., classes containing data having large differences in the amount of data.
Transfer learning (TL) [8, 9] is a family of algorithms that employs the conventional machine learning approaches with an assumption of identical distribution. Learning algorithms are moved from one domain (S) to another domain (T) to exploit and transfer insightful information. When the training data points are small, the information leveraged improvise the target domain’s learning. Transferring knowledge from one domain (S) to another domain (T) depends on the way they are connected. The stronger the association is, the greater the usability of the previous information. On the contrary, brute-force transfer in the case of poor relationships might lead to performance degradation of classifiers that negates transfer.
A general assumption of conventional machine learning approach is that training and testing dataset have similar probability distribution. Under such a presumption where a dataset is of dissimilar distribution, training data is needed to be acquired towards learning new classifiers [10]. A classifier when trained in such circumstances, it may overfit the new data which leads to poor generalization. Utilizing the knowledge from the same domain would be more effective towards regularizing the learning. Towards relaxing the similar distribution presumption of the conventional machine learning methods, transfer learning presents a family of approaches [11]. The knowledge transferred helps to improve learning in the target domain with rarity in training samples [12]. TrAdaBoost is gaining popularity for boosting by re-sampling amidst the work conducted in this area and is related to our work [13, 14]. The ability of knowledge transfers from source domain to target domain in general depends on how they relate. Earlier information will be more usable with strong relationships. Alternatively, transferring brute-force with weak relationships might deteriorate the learner’s performance and thus is termed as negative transfer [15].
In this framework, we utilized an enhanced transfer boosting approach Multisource Dynamic TrAdaBoost [16, 17] for leveraging transfer learning which considers the balance among source and target skin lesion datasets. It resolves the problem of multiclass classification using transfer learning with the same distribution of data and class unbalance amidst negative and positive instances in the source and target domain of datasets. The negative transfer problem is avoided by the approach in the datasets by transferring knowledge from multiple sources using a boosting-based framework. The approach is designed on the basis of conventional TrAdaBoost that evaluates the importance of every datapoint considered, as per the availability of number of source and target skin lesion datasets. The key contribution of proposed framework is as follows:
- A boosting-based TL framework is proposed towards transferring knowledge from multiple different sources utilizing AdaBoost, TrAdaBoost and MultiSource Dynamic TrAdaBoost (MSDTrA), for melanoma detection.
- The designed approach overcomes the challenging issues encountered during melanoma classification and trains a robust learning method with skewed dataset and scarce lesion training set.
- It improves the poor generalization ability and prevents the classifier from being overfitted with inadequate skin lesion training data.
- It avoids negative-transfer from skin lesion datasets by transferring knowledge from multiple sources using a boosting-based framework.
- It removes the biasing to the majority class and improves the prediction performance of the framework by leveraging knowledge of transfer learning from multiple sources.
- The designed framework is evaluated on seven skin lesion datasets namely, ISIC2016, ISIC2017, PH2 and HAM10000 demonstrate promising performance by alleviating negative-transfer by increasing multiple different sources.
The remaining of the work is structured as follows: Section 13.2, represents the literature based on TL methods in brief, the proposed framework is elaborately discussed in Section 13.3. Section 13.4 describes the measures for evaluating the effectiveness of framework. Experimental evaluations are represented in Section 13.5 while the work is concluded by Section 13.6.
13.2 Literature Survey
This section discusses the work conducted leveraging transfer learning to overcome the challenges confronted in datasets with skewed class distributions. One such framework is designed by Liu et al. [18], with the scarce training data towards improving the classification accuracy utilizing ensemble transfer learning framework. They first, proposed a weighted-resampling method TrResampling [19], where in each iteration TrAdaBoost algorithm is used to resample the data in the source domain and adjusts the weights for the source and the target domain.
Al-Stouhi et al. [20], tackle the problems of small dataset with imbalance in class distribution by proposing an instance transfer method with an update mechanism dependent on label, to gradually compensate for the imbalances and the issue of lack of samples. The method creates a balance with transfer learning [21] and significantly improves the classification results. This learning is known as ‘Absolute Rarity’ and is very effective in case of real-world problems. Zhang et al. [22], provided a framework which effectively builds a robust classifier that functions well in case of class imbalance. It takes the benefit of auxiliary-data using TL methods and develops a classifier that tackles with the class imbalance and gives considerable results in case of less training instances in a specific domain. This method simultaneously augments the training data and creates a balance with the unbalanced datasets. A label-dependent update mechanism is used in a novel boosting-based instance TL. Zhang et al. [13], designed an instance TL method based on multisource dynamic TrAdaBoost, considering the fact that the sample data from one domain (S) to another domain (T) have similar distribution. Knowledge is collected from different sources to avoid negative transfer. Promising classification results are achieved by the proposed algorithm using multiple source domains for leveraging transfer.
Torralba et al. [23] proposed a boosting-based multiclass classification framework which elects weak learners shared between distinct learners. They leveraged the instance based and parameter-based knowledge, transferred from different multiple sources towards boosting an individual target learner. They considered comparable amount of training instances for each task as well.
Our work overcomes the challenging issues confronted in melanoma classification as benign and malignant. The designed framework effectively deals with data scarcity and negative transfer utilizing multiple sources.
13.3 Methods and Material
Dataset
In this experiment, four benchmark skin lesion datasets namely, ISIC2016, ISIC2017, PH2, and HAM10000 are used as the source and target datasets which are available publicly. To solve the data imbalance problem for both the training and testing dataset we perform multiple augmentation operations to the images. Images were normalized, and flipped vertically and horizontally, changed the brightness, towards improving the accuracy by a significant margin [24]. Figure 13.2 shows sample images of the source and target skin lesion datasets [25]. Data augmentation process increases the size of the dataset by keeping semantic meaning of the images thus helps in reducing overfitting, the summarized detail of four target datasets is discussed in Table 13.1.
Figure 13.2 Sample skin lesion images from public dataset (a) PH2, (b) ISIC2016, (c) ISIC2017, and (d) HAM10000.
Table 13.1 Summarized detail of source and target datasets.
| Datasets | Category | Number of images | No. of classes | Index |
| PH2 [26] | Multiclass | 200 | 03 | A |
| ISIC 2016 [27] | Binary | 1,279 | 02 | B |
| ISIC 2017 [28] | Multiclass | 2,750 | 03 | C |
| HAM10000 [29] | Multiclass | 10,015 | 07 | D |
13.3.1 Proposed Methodology: Multi Source Dynamic TrAdaBoost Algorithm
The framework first utilizes TrAdaBoost with the single positive target training instance, towards utilizing the knowledge from several sources. By discarding the first half of the ensemble, TrAdaBoost degrades the performance. Due to this issue samples of source domain converge before they applied for transfer learning. In addition, relying of TrAdaBoost on single-source makes it vulnerable towards negative transfer thus, considering the co-relation among several target and source domains, we utilized ‘Multisource TrAdaBoost’ (MSTrA) [15] transfer learning method for the uneven class distribution. Ensuring that the transferred knowledge is co-related with the target domain, it neglects the impact of another source domain. We thus resolved the issue incorporated in TrAdaBoost by integrating Dynamic TrAdaBoost (DTrAdaBoost) with a dynamic-cost. However, DtrAdaBoost causes convergence of source samples prior employed for transfer learning. To overcome the aforementioned drawbacks, we then utilized multisource dynamic TrAdaBoost algorithm (MSDTrA). The method reduces the convergence-rate of weight of source instances on the basis of weak-correlation towards target domain. The MSDTrA at every step, trains the classifiers by selecting the ensemble learning of TrAdaBoost based on combining the samples of source and target. Weighted Majority Algorithm is utilized towards adjusting the source sample’s weights by reducing the weight of mis-classified source samples and keeping the present weights of correctly classified source samples. The method greatly decreases the cost of memory of unwanted data in the source domain using multiple source domains. The problem of negative transfer has been addressed by utilizing several multiple sources (skin lesion datasets) which increases the chance of importing the information from a source to the target. MSDTrA permits the training instances of source domain for participating in the learning process in every iteration, and assigns distinct weights to distinct training samples of source domain. Higher weight is assigned to the source training instance towards improving the target task learning. In general, the MSDTrA takes the benefit of significant knowledge obtained from all source domains which enhances the learning impact of target tasks. We utilized n source domains S1, S2, … Sn, n source tasks, Ta1, Ta2, …, Tan and n source training set as Da1, Da2, …, Dan, the designed framework aims to leverage the knowledge from transfer learning towards improvising the effectiveness of learning of target learner function as: ![]() The MSDTrA algorithm is more elaborately explained in pseudo code discussed in Algorithm 1 and the designed framework is depicted in Figure 13.3.
The MSDTrA algorithm is more elaborately explained in pseudo code discussed in Algorithm 1 and the designed framework is depicted in Figure 13.3.
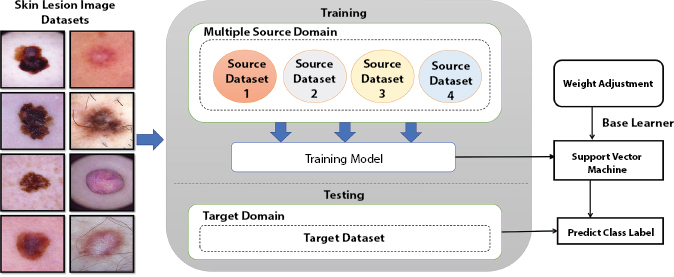
Figure 13.3 Illustration of proposed framework.
13.4 Experimental Results
Experimentations are conducted towards evaluating the effectiveness of the framework designed for detection melanoma using four benchmark skin lesion datasets namely, PH2, ISIC16, ISIC17, and HAM10000 datasets. Without losing the generalization, we considered small amount of training instances of a target skin lesion datasets while large amount of training instances of source skin lesion datasets.
13.5 Libraries Used
Following is the major open-source tools and libraries are used in conducting the experimentation. Below are the discussed libraries.
- keras library (www.keras.io)
- tensorflow library (https://www.tensorflow.org/)
- pandas library (https://pandas.pydata.org/)
- scikit-learn library (http://scikit-learn.org/)
- matplotlib library (https://matplotlib.org/)
13.6 Comparing Algorithms Based on Decision Boundaries
Figure 13.3 demonstrates the procedure of separating the samples by the algorithms. Squares represent negative instances; cross indicates positive instances belong to single source. Circles shows positive instances belong to another source. Orange colored stars (*) and crosses (+) are positive training and testing observations in the target domain correspondingly. Figure 13.4(a) represents overfitting of target data with poor generalization by the AdaBoost. Figure 13.4(b) demonstrates the decision boundaries achieved by TrAdaBoost when two sources are employed jointly. Figure 13.4(c) represents improved decision boundaries obtained by MSDTrA where every source separately combined with the target.
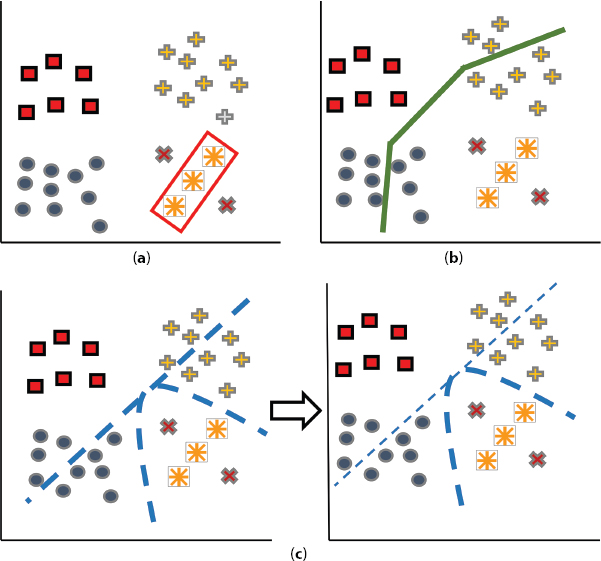
Figure 13.4 Representing decision boundary amid negative and positive instances in the target domain computed by (a) AdaBoost, (b) TrAdaBoost and (c) MSDTrA. Orange colored stars and crosses demonstrate positive instances of the target, dashed lines show candidate- decision boundary while solid line represents learned boundary.
13.7 Evaluating Results
The performance of MSDTrA is evaluated on four pigmented lesion datasets. The datasets employed comprise of small amount of training observations with respect to target data and abundant training instances corresponding to source datasets. We conducted a fair comparison of employed methods namely AdaBoost, TrAdaBoost with the MSDTrA to test its generalized characteristics. Liner Support Vector Machine is used as a base learner towards developing a weak learner for every experimentation. The performance of methods is evaluated using the receiver operating curve (ROC) and area under the ROC curve (AUCROC). Since the number of positive samples for training was very limited, image augmentation was performed using different transformations to increase the number of images. We varied the source domains from A-D towards investigating the learner’s performance corresponding to the domain’s variability.
The amount of positive instances considered for one data source is 5,000, while the negative instances of both the domains are drawn at random from the augmented dataset. The amount of negative training instances in the target dataset is set to 20% of the source domain dataset. Similarly, the amount of negative test instances in the target dataset is set to 500. For every target domain, the learner’s performance is assessed over the six random combinations of source domains categories. With given source and target domain, the performance of learner is achieved by taking the average over fifty trials of experimentations while the overall learner’s performance is average on four target sets.
Figure 13.5 represents visual illustration of comparison of AdaBoost, TrAdaBoost, and MSDTrA based on AUCROC with distinct amount of positive target training instances as 1, 5, 15, 50 and source domain N = 1, 2, 3, 5. Figure 13.5(a) considers N = 3 and demonstrates the behavior of methods as the number of positive training instances increases. Since AdaBoost doesn’t perform knowledge transfer from the source, its performance is dependent on number of positive training instances. TrAdaBoost integrates the three sources into one and improvised the performance utilizing TL mechanism. By transferring knowledge from multiple single domains, MultiSource Dynamic TrAdaBoost demonstrates significant improvement in classification accuracy, even with small training sample. TrAdaBoost integrates the three sources into one, improves on AdaBoost because of transfer learning mechanism. MSDTrA shows significant improvements in accuracy even with small amount of positive training instances by incorporating knowledge transfer from multiple single domains.
Figure 13.5(b) considers N = 1, which demonstrate that TrAdaBoost reduces to AdaBoost and thus, showing similar performances. While TrAdaBoost outperforms MultiSource Dynamic TrAdaBoost (MSDTrA) when the number of positive training instances is less and performs poor with large number of positive training instances. Figure 13.5(c) considers number of positive training instances = 1, and proves that with increase in source domain, AUCROC of MultiSource Dynamic TrAdaBoost increases with respect to decrease in standard deviations. This indicates an enhance performance in terms of consistency and accuracy. Figure 13.5(d) plots the average training-time with respect to experimentation conducted against number of positive training instances and N of entire methods.
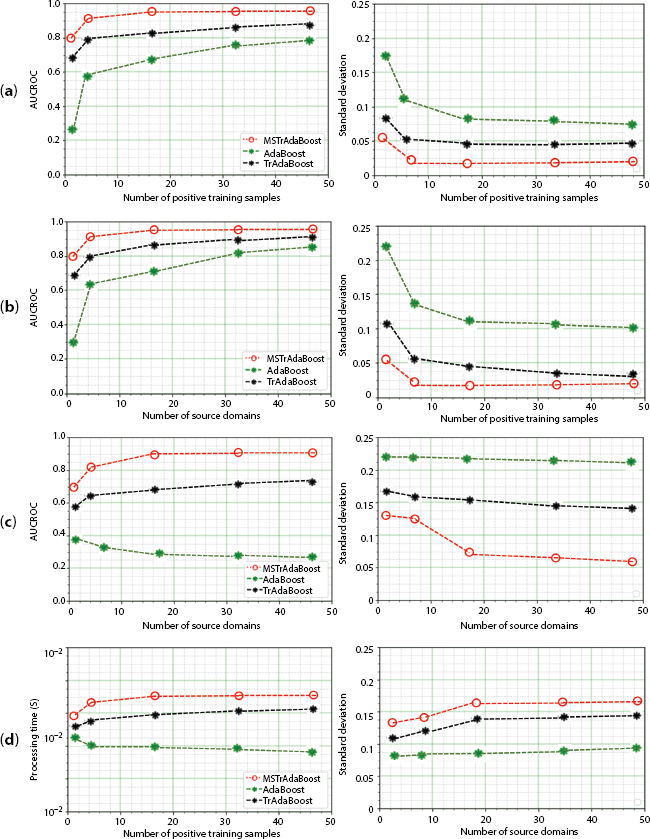
Figure 13.5 Performance comparison analysis. (a) AUCROC respective to standard deviation against amount of positive training instances with N = 3 sources. (b) AUCROC respective to standard deviation against amount of positive training instances with . (c) AUCROC respective to standard deviation against amount of positive training instances with N = 1. (d) Execution time against amount of positive training instances with N = 2 (upper), and N = 1 (lower).
13.8 Conclusion
The relationship among source and target domain affects the impact of transfer learning. Instead of improving the process of learning, leveraging the brute-force poorly from the source with respect to the target might degrade the performance of a classifier. We reduced the negative-transfer by importing of knowledge from multiple different sources for increasing the chance of searching a source corresponding to the target. In this work, MultiSource Dynamic TrAdaBoost (MSDTrA) an instance transfer-learning approach is designed for the classification of imbalanced class distribution in pigmented skin lesion datasets towards melanoma detection. The proposed framework integrates the knowledge obtained from different source domains and resolves the problem of multiclass classification using transfer learning with the same distribution of data and class unbalance among negative and positive instances of datasets. The proposed method averts the effect of negative-transfer and skewed class distribution in skin lesion datasets by incorporating multiple sources. The experimental results and theoretical analysis illustrate that the proposed method provides promising performance of classification than the several conventional methods.
References
1. Mendonca, T., Ferreira, P.M., Marques, J.S., Marcal, A.R.S., Rozeira, J., PH2–A dermoscopic image database for research and benchmarking, in: Conference proceedings: Annual International Conference of the IEEE Engineering in Medicine and Biology Society, vol. 2013, pp. 5437–40, 2013.
2. Hosny, K.M., Kassem, M.A., Fouad, M.M., Classification of Skin Lesions into Seven Classes Using Transfer Learning with AlexNet. J. Digit. Imaging, 33, 5, 1325–1334, 2020.
3. Li, Y. and Shen, L., Skin lesion analysis towards melanoma detection using deep learning network. Sensors, 18, 2, 556, 2018.
4. Amelard, R., Glaister, J., Wong, A., Clausi, D.A., High-Level Intuitive Features (HLIFs) for intuitive skin lesion description. IEEE Trans. Biomed. Eng., 62, 3, 820–831, 2015.
5. Noar, S.M., Leas, E., Althouse, B.M., Dredze, M., Kelley, D., Ayers, J.W., Can a selfie promote public engagement with skin cancer? Preventive Med., 111, November, 280–283, 2018.
6. A.C. Society, Colorectal cancer facts & figures 2014–2016, American Cancer Society, Atlanta, Georgia, 2014.
7. Al-Stouhi, S., Learning with an insufficient supply of data via knowledge transfer and sharing. Wayne State University. ProQuest Dissertations and Thesis, p. 1–148, 2013, [Online]. https://manchester.idm.oclc.org/
8. Pan, S.J., Transfer learning, in: Data Classification: Algorithms and Applications, pp. 537–570, 2014.
9. Singh, L., Janghel, R.R., Sahu, S.P., TrCSVM: A novel approach for the classification of melanoma skin cancer using transfer learning. Data Technol. Appl., 55, 1, 1–18, 2020.
10. Bang, S.H., Ak, R., Narayanan, A., Lee, Y.T., Cho, H., A survey on knowledge transfer for manufacturing data analytics. Comput. Ind., 104, 116–130, 2019.
11. Yuan, Z., Bao, D., Chen, Z., Liu, M., Integrated Transfer Learning Algorithm Using Multi-source TrAdaBoost for Unbalanced Samples Classification, in: International Conference on Computing Intelligence and Information System, (CIIS), pp. 188–195, 2017.
12. Yuhu, C., Ge, C.A.O., Xuesong, W., Jie, P.A.N., Weighted Multi-source TrAdaBoost *. 22, 3, 505–10, 2013.
13. Zhang, Q., Li, H., Zhang, Y., Li, M., Instance Transfer Learning with Multisource Dynamic TrAdaBoost Qian. Sci. World J., 10, 1–8, 2014.
14. Liu, X., Wang, G., Cai, Z., Zhang, H., A multiboosting based transfer learning algorithm. J. Adv. Comput. Intell. Intell. Inform., 19, 3, 381–388, 2015.
15. Yao, Y. and Doretto, G., Boosting for transfer learning with multiple sources, in: Proceedings of the IEEE Computer Society Conference on Computer Vision and Pattern Recognition, pp. 1855–1862, 2010.
16. Eaton, E. and Desjardins, M., Set-based boosting for instance-level transfer, in: IEEE International Conference on Data Mining (ICDM), pp. 422–428, 2009.
17. Wenyuan Dai, Y.Y., Yang, Q., Xue, G.-R., Boosting for Transfer Learning, in: Proceedings of the 24th International Conference on Machine Learning, pp. 193–200, 2007.
18. Liu, X., Liu, Z., Wang, G., Cai, Z., Zhang, H., Ensemble Transfer Learning Algorithm. IEEE Access, 6, 2389–2396, 2017.
19. Liu, X., Liu, Z., Wang, G., Cai, Z., Zhang, H., A weighted-resampling based transfer learning algorithm. Proceedings of the International Joint Conference on Neural Networks, May, vol. 2017, pp. 185–190, 2017.
20. Al-Stouhi, S. and Reddy, C.K., Transfer learning for class imbalance problems with inadequate data. Knowl. Inf. Syst., 48, 1, 201–228, 2016.
21. Weiss, K., Khoshgoftaar, T.M., Wang, D., A survey of transfer learning. J. Big Data, 3, 1–40, 2016.
22. Zhang, X., Zhuang, Y., Wang, W., Pedrycz, W., Transfer boosting with synthetic instances for class imbalanced object recognition. IEEE Trans. Cybern., 48, 1, 357–370, 2018.
23. Torralba, K., Murphy, P., Freeman, W.T., Sharing visual features for multiclass and multiview object detection. IEEE Trans. Pattern Anal. Mach. Intell., 29, 5, 854–869, 2007.
24. Pham, T.-C., Luong, C.-M. Visani, M., Hoang, V.-D., Deep CNN and data augmentation for skin lesion classification. In Asian Conference on Intelligent Information and Database Systems, Springer, Cham, pp. 573–582, 2018.
25. Tschandl, P., Rosendahl, C., Kittler, H., The HAM10000 dataset, a large collection of multi-source dermatoscopic images of common pigmented skin lesions. Scientific data, 5, 1, 1–9, 2018.
26. Teresa Mendonca, J.R., Ferreira, P.M., Marcal, A.R.S., Barata, C., Marques, J.S., Rocha, J., PH2: A Public Database for the Analysis of Dermoscopic Images, in: Dermoscopy Image Analysis, pp. 419–439, April 2015.
27. Gutman, D., Codella, N.C.F., Celebi, E., Helba, B., Marchetti, M., Mishra, N., Skin Lesion Analysis toward Melanoma Detection: A Challenge at the International Symposium on Biomedical Imaging (ISBI) 2016, hosted by the International Skin Imaging Collaboration (ISIC). In [Online]. Available: https://arxiv.org/abs/1605.01397.
28. Noel, K.L., Codella, C.F., Gutman, D.M., Celebi, E., Helba, B., Marchetti, M.A., Dusza, S.W., Kalloo, A., Nabin Mishra, A.H., Kittler, H., Skin lesion analysis toward melanoma detection: A challenge at the 2017 International Symposium on Biomedical Imaging (ISBI), hosted by the International Skin Imaging Collaboration (ISIC). Proceedings—International Symposium on Biomedical Imaging, 2018-April, pp. 168–172, 2018.
29. Tschandl, P., Rosendahl, C., Kittler, H., Data descriptor: The HAM10000 dataset, a large collection of multi-source dermatoscopic images of common pigmented skin lesions. Sci. Data, 5, 1–9, 2018.
- *Corresponding author: [email protected]
