Tab. 6.11: J48 algorithm experiment result.
Note:* denotes a dataset with missing values, and CCI/DOA denote classified correctly (instances) and degree of accuracy, respectively.
Tab. 6.12: DFGHR algorithm experiment result.
Note:* denotes a dataset with missing values, and CCI/DOA denote classified correctly (instances) and degree of accuracy, respectively.
6.6.1Question description
First, we consider the verification of chemical molecular structure identification results. Data mining and knowledge representation of chemical molecular information is an important field of life sciences. By analysing the chemical molecular structure, we can abstract a given substance to an undirected graph, where the vertices in the graph denote the atoms and the relations between the vertices represent the chemical bonds between the atoms. A double sparse relational learning algorithm based on multiple kernel learning [2] has been used to test chemical information datasets, and experimental results show that the prediction accuracy is obviously improved, with fewer rule sets and more direct explanations. Zhao XF et al. [58] designed the OpenMolGRID system, which is used to mine chemical information in the grid environment for the design and construction of molecules. Qi et al. [59] proposed a two-layer classifier to identify the Escherichia coli promoter in DNA. The first-level classifier uses three Bayesian neural networks to learn three different feature sets, and the second layer is combined with the output of the first layer to give the final results. The method of Borgelt et al. [60] embeds the generated molecular fragments in parallel into all suitable molecular structures and prunes the search tree to help distinguish between different classes based on local atomic and bond sequences. Machine learning methods help chemists in two ways: finding frequently occurring chemical molecular fragments and finding fragments in the database that are frequent and infrequent in other parts of the database, which allows the database to be divided into active molecules and non-active molecules, as shown in the following example.


For the compounds shown in Fig. 6.14, two are active and two are inactive. The task of learning is to find a model that can distinguish active from non-active molecules. This type of learning task is important in computational chemistry, where it is usually called structure activity relationship prediction [1] and can be used for the design and development of drugs and toxicity detection.



The pattern found in Fig. 6.13 is called a structural alarm and is used to separate active molecules from inactive molecules. Because the structure warning is a substructure (sub-graph), it is compatible with the active molecule, and there is no match with the non-active molecules. At the same time, the structural alarm is easy to understand and provides useful insights into the factors that determine molecular activity.
The DF map constructed by DFGHR is compressed using the Subdue method [57]. Because it can find common sub-graph patterns in the large image, we iterate the model to compress the original image until it can no longer be compressed. This produces a hierarchical conceptual clustering of the input data. In the i th iteration, the best sub-graph is used to compress the input graph and introduce the new node into the graph for the next iteration, as shown in Fig. 6.14.
Figure 6.16 hierarchy discovery process can be compressed as shown in Fig. 6.15.

6.6.2Sample analysis
We combined the DFGHR and DFLR algorithms to construct a DF graph and extract rules from the “promoter” dataset in the UCI knowledge base (http://www.ics.uci.edu/~mlearn/MLRepository.html). The input properties are 57 ordered DNA nucleotides (A, G, T, C), with a total sample number of 106 [53 positive cases (promoter sequence samples), 53 negative examples (non-promoter sequence samples)]. Each nucleotide sequence is aligned with a reference point, so that the n th attribute corresponds to the n th nucleotide. If the data are not aligned, standard classifiers such as C4.5 cannot handle the problem. In the form of a graph, the data need not be aligned with the reference points.
In this section, each sequence is transformed into a representation of a graph, as shown in Fig. 6.17. Each element is assumed to interact with at most 10 elements. Therefore, each sequence obtains 57 nodes and 515 connections.
After using the DFGHR algorithm to produce a sub-graph model of the promoter dataset, the DFLR algorithm was used to obtain the following classification rules: where y represents the abstracted sub-structure, n represents the number of models abstracted, + represents a promoter, and – represents a non-promoter.

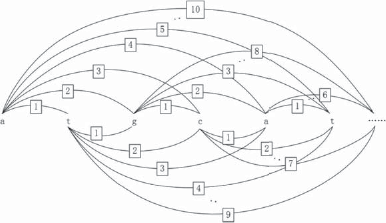

The resulting rule size is compared with those of the C4.5 and MofN algorithms in Fig. 6.18.
Intelligibility reflects the extent to which the rule is understood by the user. Any increase in the rule number and the antecedent number of each rule will increase the difficulty of understanding the rules. From the above graph, we can see that the rules obtained by DFLR are simpler to understand than those obtained by C4.5 and MofN. It is also necessary to consider whether the rules have significance. Of the six rules obtained by DFLR, the sixth rule covers counterexamples. This has too many complex antecedents, and can be omitted. That is, the samples covered by the first five rules are positive, otherwise they are negative.
6.7Summary
Relationship learning is a learning paradigm in the field of machine learning that considers knowledge representation to obtain specific information in relational data, and then uses the knowledge obtained to reason, predict, and classify.
It is difficult to deal with uncertain information, such as the bias in data caused by the subjectivity of an expert system, or the absence or fuzziness of data. Dynamic fuzzy theory can effectively express this kind of data.
This chapter has introduced the theory of DFD into relational learning. The main results include the following:
(1)Based on DFL and a dynamic fuzzy matrix, we proposed a DFL learning algorithm and dynamic fuzzy relationship matrix hierarchy learning algorithm, and verified the effectiveness of the algorithms.
(2)On the basis of dynamic fuzzy sets and the dynamic fuzzy production, combined with the C4.5 algorithm and a decision tree, a dynamic fuzzy tree hierarchical relationship learning algorithm was proposed, and samples combined with real data were analysed.
(3)Based on dynamic fuzzy graph theory, we proposed a dynamic fuzzy direct acyclic graph hierarchy analysis, and presented a dynamic fuzzy HRL algorithm that was compared with the J48 algorithm.
(4)By combining the DFGHR and DFLR algorithms, a dataset of chemical molecules with missing data was constructed and its rules extracted. Compared with the conventional method, our algorithm is more general.
References
[1]Raedt LD. Logical and relational learning. New York, USA, Springer-Verlag Inc., 2008.
[2]Han YJ, Wang Y YJ. A Bi-sparse relational learning algorithm based on multiple kernel learning. Journal of Computer Research and Development, 2010, 47(8): 1400–1406.
[3]Sato T, Kameya Y. PRISM: A symbolic-statistical modeling language. In Proceedings of the 15th International Joint Conference on Artificial Intelligence (IJCAI-97). San Francisco, Morgan Kaufmann, 1997, 1330–1339.
[4]Muggleton S. Stochastic logic programs. In Proceedings of the 5th International Workshop on Inductive Logic Programming. Amsterdam, USA, IOS Press, 1996, 254–264.
[5]Kramer S. Structural regression trees. In Proceedings of the 13th National Conference on Artificial Intelligence. Menlo Park, CA, AAAI Press, 1996, 812–819.
[6]Blckeel H A, Raedt L De. Top-down induction of first-order logical decision trees. Artificial Intelligence, 1998, 101(1/2): 285–297.
[7]Knobbe A J, Siebes A, Der Wallen D Van. Multi-relational decision tree induction. In Proceedings of the 3rd European Conference on Principles and Practice of KDD, Berlin, Germany, Springer, 1999, 378–383.
[8]Ngo L, Haddawy P. Answering queries from context-sensitive probabilistic knowledge bases. Theoretical Computer Science, 1997, 171(1/2): 147–177.
[9]Kersting K, Raedt L De. Adaptive Bayesian logic programs. In Proceedings of the 11th Conference on Inductive Logic Programming. Berlin, Germany, Springer, 2001, 104–117.
[10]Muggleton S. Learning stochastic logic programs. In Proceedings of the AAAI2000 Workshop on Learning Statistical Models from Relational Data. Menlo Park, CA, AAAI Press, 2000, 19–26.
[11]Cussens J. Parameter estimation in stochastic logic programs. Machine Learning, 2001, 44(3): 245–271.
[12]Muggleton S. Learning structure and parameters of stochastic logic programs //LNCS 2583: Proceedings of the 12th International Conference on Inductive Logic Programming. Berlin, Germany, Springer, 2002, 198–206.
[13]Neville J, Jensen D, Friedland L, et al. Learning relational probability trees. In Proceedings of the 9th International Conference on Knowledge Discovery & Data Mining. New York, USA, ACM, 2003, 9–14.
[14]Bernstein A, Clearwater S, Provost F. The relational vector-space model and industry classification. In Proceedings of IJCAI2003, Workshop on Learning Statistical Models from Relational Data. San Francisco, USA, Morgan Kaufmann, 2003, 8–18.
[15]Angelopoulos N, Cussens J. On the implementation of MCMC proposals over stochastic logic programs. In Proceedings of Colloquium on Implementation of Constraint and Logic Programming Systems. Satellite Workshop to ICLP’04. Berlin, Germany, Springer, 2004.
[16]Taskar B, Abbeel P, Koller D. Discriminative probabilistic models for relational data. In Proceedings of the 18th Conference on Uncertainty in Artificial Intelligence. San Francisco, USA, Morgan Kaufmann, 2002, 485–492.
[17]Richardson M, Domingos P. Markov logic networks. Seattle, WA, Department of Computer Science and Engineering, University of Washington, 2004.
[18]Anderson C, Domingos P, Weld DS. Relational Markov models and their application to adaptive Web navigation. In Proceedings of the 8th International Conference on Knowledge Discovery and Data Mining (KDD-02). New York, ACM, 2002, 143–152.
[19]Kersting K, Raiko T, Kramer S, et al. Towards discovering structural signatures of protein folds based on logical hidden Markov models. In Proceedings of the 8th Pacific Symposium on Biocomputing, 2003, 192–203.
[20]Davis J, Burnside E, Dutra IC, et al. An integrated approach to learning Bayesian networks of rules. Proceedings of the 16th European Conference on Machine Learning. Berlin, Germany, Springer, 2005, 84–95.
[21]Landwehr N, Kersting K, Raedt L De. nFOIL: Integrating naive Bayes and FOIL. In Proceedings of AAAI. Berlin, Germany, Springer, 2005, 795–800.
[22]Sato T. A generic approach to EM learning for symbolic-statistical models. In Proceedings of ICML05 Workshop on Learning Language in Logic. New York, USA, ACM, 2005,1130–1200.
[23]Izumi Y, Kameya Y, Sato T. Parallel EM learning for symbolic-statistical models. In Proceedings of the International Workshop on Data-Mining and Statistical Science. Sapporo, Japan, 2006, 133–140.
[24]Xu Z, Tresp V, Yu K, et al. Infinite hidden relational models. In Proceedings of the 22nd Conference on Uncertainty in Artificial Intelligence. Arlington, Virginia, AUAI Press, 2006.
[25]Singla P, Domingos P. Entity resolution with Markov logic. In Proceedings of the 6th International Conference on Data Mining. Los Alamitos, CA, IEEE Computer Society, 2006, 572–582.
[26]Kersting K, Raedt LD, Raiko T. Logical hidden Markov models. Journal of Artificial Intelligence Research, 2006, 25: 425–456.
[27]Chen J, Muggleton SH. A revised comparison of Bayesian logic programs and stochastic logic programs. In Proceedings of the 16th International Conference on Inductive Logic Programming. Berlin, Germany, Springer, 2006.
[28]Singla P, Domingos P. Markov logic in infinite domains. In Proceedings of the 23rd Conference on Uncertainty in Artificial Intelligence. Arlington, VA, AUAI Press, 2007, 368–375.
[29]Wang J, Domingos P. Hybrid Markov logic networks. In Proceedings of the 23rd AAAI Conference on Artificial Intelligence. Menlo Park, CA, AAAI Press, 2008, 1106–1111.
[30]Petr B, Jiri K, Katsumi I. Grammatical concept representation for randomised optimization algorithms in relational learning. In Proceedings of the 9th International Conference on Intelligent Systems Design and Applications, 2009, 156, 1450–1455.
[31]Zhu M. DC Proposal: Ontology learning from noisy linked data. USA, Springer-Verlag, 2011, 2(4): 373–380.
[32]Bengio Y. Learning Deep Architectures for AI[J]. Foundations & Trends® in Machine Learning, 2009, 2(1): 1–127.
[33]Hinton GE, Osindero S, Teh YW. A fast learning algorithm for deep belief nets. Neural Computation, 2014, 18(7): 1527–1554.
[34]Bengio Y, Lamblin P, Dan P, et al. Greedy layer-wise training of deep networks[C]. International Conference on Neural Information Processing Systems. MIT Press, 2006:153–160. Bengio Y, Lamblin P, Popovici D, Larochelle H. Greedy layer-wise training of deep networks. In Scholkopf B, Platt J, Hoffman T (Eds.). In Advances in neural information processing systems 19, 2006, 153–160.
[35]Erhan D, Manzagol PA, Bengio Y, Bengio S, Vincent P. The difficulty of training deep architectures and the effect of unsupervised pre-training. In Proceedings of the Twelfth International Conference on Artificial Intelligence and Statistics, 2009, 153–160.
[36]Hugo L, Yoshua B, Jerme L, et al. Exploring strategies for training deep neural networks. Journal of Machine Learning Research, 2009, 10: 1–40.
[37]Ranzato M, Boureau YL, LeCun Y. Sparse feature learning for deep belief networks. In Platt J, Koller D, Singer Y, & Roweis S (Eds.), Cambridge, MA, In Advances in Neural Information Processing System, 2007, 1185–1192.
[38]Salakhutdinov R, Hinton GE. Deep Boltzmann machines. Journal of Machine Learning Research, 2009, 5(2): 1967–2006.
[39]Jason W, Frederic R, Ronan C. Deep learning via semi-supervised embedding. In Proceedings of the 25th International Conference on Machine Learning, 2008, 307, 1168–1175.
[40]Ranzato, Marc A, Huang, et al. Unsupervised learning of invariant feature hierarchies with applications to object recognition. In Proceedings of the Computer Vision and Pattern Recognition Conference, 2007.
[41]Lee H, Grosse R, Ranganath R, et al. Convolutional deep belief networks for scalable unsupervised learning of hierarchical representations. In Proceedings of the 26th International Conference on Machine Learning, 2009.
[42]Sun M, Chen B, Zhou MT. Domain relation learning from events in log ontologies based on hierarchy association rules. Application Research of Computers, 2009, 26(10): 3683–3686.
[43]Cook D, Holder L. Graph-based data mining. IEEE Intelligent Systems, 2000, 15(2): 32–41.
[44]Yoshida K, Motoda H, Indurkhya N. Graph-based induction as a unified learning framework. Journal of Applied Intelligence, 1994, 4: 297–328.
[45]Li FZ, et al. Dynamic fuzzy set and application. Kunming, China, Yunnan Science & Technology Press, 1997.
[46]Li FZ, Zhu WH. Dynamic fuzzy logic and Application. Kunming, China, Yunnan Science & Technology Press, 1997.
[47]Li FZ, Liu GQ, She YM. An introduction to dynamic fuzzy logic. Kunming, China, Yunnan Science & Technology Press, 2005.
[48]Li FZ. Dynamic fuzzy logic and its application. America, Nova Science Publishers, 2008.
[49]Li FZ. Research on a dynamic fuzzy relation data model. Journal of Chinese Mini-Micro Computer Systems, 2002, 23(9): 1107–1109.
[50]Zhang J. Dynamic fuzzy machine learning model and its applications. Master’s Thesis, Soochow University, Suzhou, China, 2007.
[51]Mitchell, TM. Machine learning. NY, USA, McGraw-Hill, 1997.
[52]Henri P, Gilles R, Mathieu S. Enriching relational learning with fuzzy predicates. In Proceedings of PKDD, 2003.
[53]Sheng Z, Xie SQ, Pan CY. Probability theory and mathematical statistics. Beijing, China, Higher Education Press, 2001: 176–207.
[54]Delgado M, Sanchez D, and Vila MA. Fuzzy cardinality based evaluation of quantified sentences. International Journal of Approximate Reasoning, 2000, 23(1): 23–66.
[55]Frederic S, David M. Using kernel basis with relevance vector machine for feature selection. In Proceedings of ICANN, 2009, 5769: 255–264.
[56]Kahn AB. Topological sorting of large networks. Communications of the ACM, 1962, 5(11): 558–562.
[57]Maran U, Sild S, Kahn I, Takkis K. Mining of the chemical information in GRID environment. Future Generation Computer Systems, 2007, 23: 76–83.
[58]Qicheng MA, Jason TL. Wang. Biological data mining using bayesian neural networks: a case study[J]. International Journal on Artificial Intelligence Tools, 1999, 8(04).
[59]Borgelt C, Berthold MR. Mining molecular fragments: Finding relevant substructures of molecules. In Proceeding of the 2002 international conference on data mining, 2002, 2: 211–218.
[60]Tammy RR, Nir S, Nahum K. On symmetry, perspectivity, and level set based segmentation. IEEE transactions on pattern analysis and machine intelligence, 2009, 31(8): 1458–1471.
[61]Holder L, Cook DJ, Coble J, and Mukherjee M. Graph-based relational learning with application to security. Fundamental Informaticae Special Issue on Mining Graphs, Trees and Sequences, 2005, 66(1–2): 83–101.
[62]Hsu W, Joehanes R. Relational decision networks. In Proceedings of the ICML-2004 Workshop on Statistical Relational Learning and Connections to Other Fields, 2004, 61–67.
[63]Li FF, Rob F, Antonio T. Recognizing and Learning Object Categories. In Proceedings of ICCV, 2009.
[64]David GL. Distinctive image features from scale-invariant keypoints. International Journal of Computer Vision, 2004, 60(2), 91–11.
[65]Ontanon S, Plaza E. Similarity measures over refinement graphs. Machine Learning, 2012, 87: 57–92.
[66]Rettinger A, Nickles M, Tresp V. Statistical relational learning of trust. Machine Learning, 2011, 82: 191–209.
[67]Mustafa D. The order-theoretic duality and relations between partial metrics and local equalities. Fuzzy Sets and Systems, 2012, 192: 5–57.
[68]Ignjatović J, et al. Weakly linear systems of fuzzy relation inequalities: The heterogeneous case. Fuzzy Sets and Systems, 2011, 199(24): 64–91.


