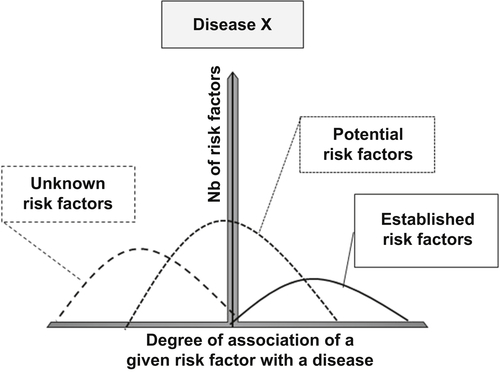
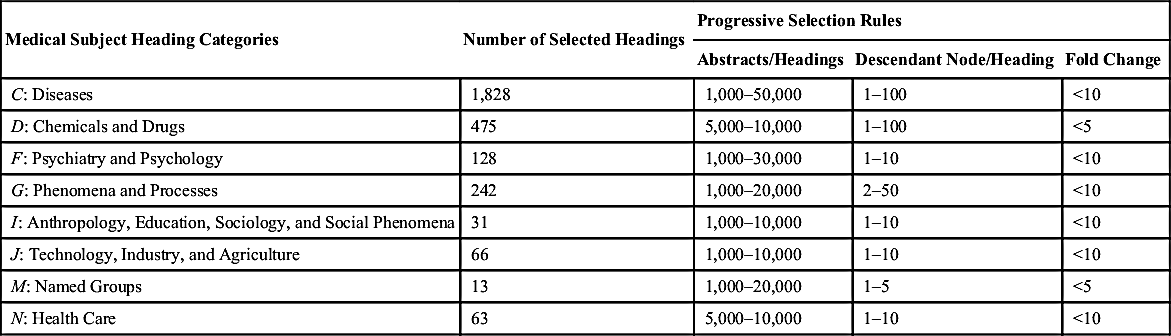
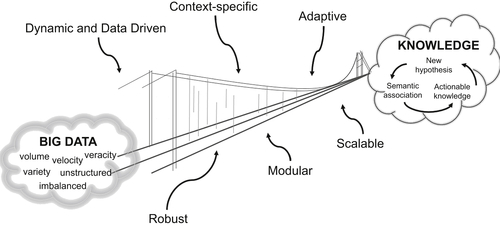
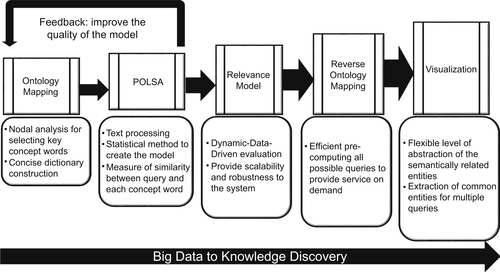
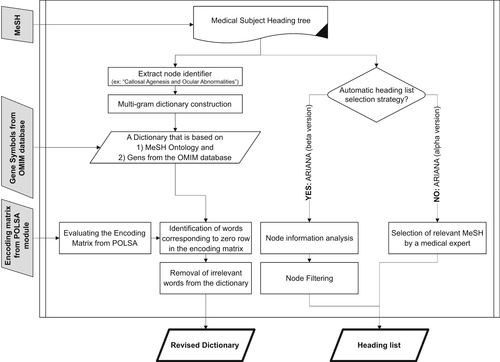
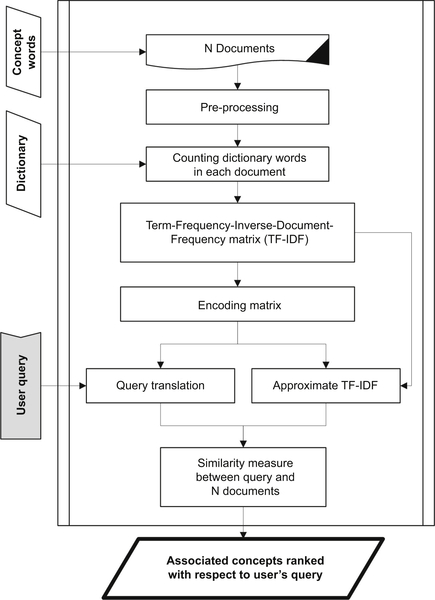
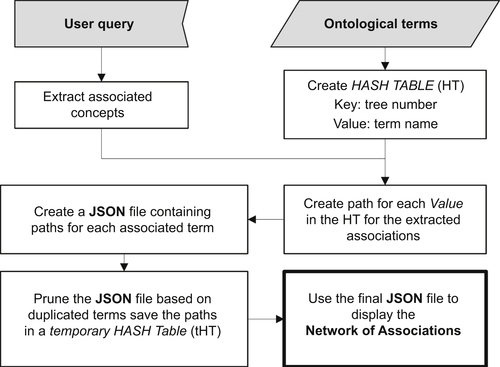
Abedi V, Yeasin M, Zand R. ARIANA: adaptive robust and integrative analysis for finding novel associations. In: The 2014 International Conference on Advances in Big Data Analytics. Las Vegas, NV. 2014.
Abedi V, Zand R, Yeasin M, Faisal F.E. An automated framework for hypotheses generation using literature. BioData Mining. 2012;5:13. doi: 10.1186/1756-0381-5-13.
Altman R.B, Bergman C.M, Blake J, Blaschke C, Cohen A, Gannon F, Grivell L, Hahn U, Hersh W, Hirschman L, Jensen L.J, Krallinger M, Mons B, O’Donoghue S.I, Peitsch M.C, Rebholz-Schuhmann D, Shatkay H, Valencia A. Text mining for biology–the way forward: opinions from leading scientists. Genome Biology. 2008;9(Suppl. 2):S7. doi: 10.1186/gb-2008-9-s2-s7.
Ashburner M, Ball C.A, Blake J.A, Botstein D, Butler H, Cherry J.M, Davis A.P, Dolinski K, Dwight S.S, Eppig J.T, Harris M.A, Hill D.P, Issel-Tarver L, Kasarskis A, Lewis S, Matese J.C, Richardson J.E, Ringwald M, Rubin G.M, Sherlock G. Gene ontology: tool for the unification of biology. The gene ontology consortium. Nature Genetics. 2000;25:25–29. doi: 10.1038/75556.Gene.
Berman J.J. Pathology abbreviated: a long review of short terms. Archives of Pathology & Laboratory Medicine. 2004;128:347–352. doi: 10.1043/1543-2165(2004)128<347:PAALRO>2.0.CO;2.
Berry M.P.R, Graham C.M, McNab F.W, Xu Z, Bloch S.A.A, Oni T, Wilkinson K.A, Banchereau R, Skinner J, Wilkinson R.J, Quinn C, Blankenship D, Dhawan R, Cush J.J, Mejias A, Ramilo O, Kon O.M, Pascual V, Banchereau J, Chaussabel D, O’Garra A. An interferon-inducible neutrophil-driven blood transcriptional signature in human tuberculosis. Nature. 2010;466:973–977. doi: 10.1542/peds.2011-2107LLLL.
Berry W.M, Browne M. Understanding Search Engines: Mathematical Modeling and Text Retrieval. Philadelphia, PA, USA: Society for Industrial and Applied Mathematics Philadelphia; 1999.
Brettner A, Heitzman E.R, Woodin W.G. Pulmonary complications of drug therapy. Radiology. 1970;96:31–38. doi: 10.1148/96.1.31.
Brinckerhoff C.E, Matrisian L.M. Matrix metalloproteinases: a tail of a frog that became a prince. Nature Reviews Molecular Cell Biology. 2002;3:207–214. doi: 10.1038/nrm763.
Cockcroft A, Masisi M, Thabane L, Andersson N. Science Communication. legislators learning to interpret evidence for policy. Science. 2014;345:1244–1245. doi: 10.1126/science.1256911.
Cockersole F.J, Park W.W. Hexamethonium lung; report of a case associated with pregnancy. Journal of Obstetrics and Gynaecology of the British Empire. 1956;63:728–734.
Collier N, Doan S, Kawazoe A, Goodwin R.M, Conway M, Tateno Y, Ngo Q.-H, Dien D, Kawtrakul A, Takeuchi K, Shigematsu M, Taniguchi K. BioCaster: detecting public health rumors with a Web-based text mining system. Bioinformatics. 2008;24:2940–2941. doi: 10.1093/bioinformatics/btn534.
Davidson J.M. Biochemistry and turnover of lung interstitium. European Respiratory Journal. 1990;3:1048–1063.
Elkington P.T, Ugarte-Gil C.A, Friedland J.S. Matrix metalloproteinases in tuberculosis. European Respiratory Journal. 2011;38:456–464. doi: 10.1183/09031936.00015411.
Franceschini A, Szklarczyk D, Frankild S, Kuhn M, Simonovic M, Roth A, Lin J, Minguez P, Bork P, von Mering C, Jensen L.J. STRING v9.1: protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Research. 2013;41:D808–D815. doi: 10.1093/nar/gks1094.
Garcia P, Armstrong R, Zaman M.H. Models of education in medicine, public health, and engineering. Science. 2014;345:1281–1283. doi: 10.1126/science.1258782.
Hirschman L, Morgan A.A, Yeh A.S. Rutabaga by any other name: extracting biological names. Journal of Biomedical Informatics. 2002;35:247–259.
Internal Investigative Committee Membership. Report of Internal Investigation into the Death of a Volunteer Research Subject [Online]. 2001 Available from. http://www.hopkinsmedicine.org/press/2001/july/report_of_internal_investigation.htm.
Kim J.-J, Pezik P, Rebholz-Schuhmann D. MedEvi: retrieving textual evidence of relations between biomedical concepts from Medline. Bioinformatics. 2008;24:1410–1412. doi: 10.1093/bioinformatics/btn117.
Kirkland K.E, Kirkland K, Many W.J, Smitherman T.A. Headache among patients with HIV disease: prevalence, characteristics, and associations. Headache. 2012;52:455–466. doi: 10.1111/j.1526-4610.2011.02025.x.
Landauer T.K, Dumais S.T. A solution to Plato’s problem: the latent semantic analysis theory of acquisition, induction, and representation of knowledge. Psychological Review. 1997;104:211–240. doi: 10.1037/0033-295X.104.2.211.
Landauer T.K, Laham D, Derr M. From paragraph to graph: latent semantic analysis for information visualization. Proceedings of the National Academy of Sciences. 2004;101:5214–5219. doi: 10.1073/pnas.0400341101.
Lu Z. PubMed and beyond: a survey of web tools for searching biomedical literature. Database (Oxford). 2011:baq036. doi: 10.1093/database/baq036.
Malaia L.T, Shalimov A.A, Dushanin S.A, Liashenko M.M, Zverev V.V. Catheterization of veins and selective angiopulmonography in comparison with several indices of the functional state of the external respiratory apparatus and blood circulation during chronic lung diseases. Kardiologiia. 1967;7:112–119.
Mehra S, Pahar B, Dutta N.K, Conerly C.N, Philippi-Falkenstein K, Alvarez X, Kaushal D. Transcriptional reprogramming in nonhuman primate (Rhesus Macaque) tuberculosis granulomas. PLoS One. 2010;5 doi: 10.1371/journal.pone.0012266.
Nasukawa T, Yi J. Sentiment analysis. In: Proceedings of the International Conference on Knowledge Capture - K-cap ‘03. New York, USA: ACM Press; 2003:70. doi: 10.1145/945645.945658.
National Center for Biotechnology Information, n.d. Phenotype-genotype Integrator [Online]. Available from: http://www.ncbi.nlm.nih.gov/gap/phegeni.
Nishida Y, Tandai-Hiruma M, Kemuriyama T, Hagisawa K. Long-term blood pressure control: is there a set-point in the brain? Journal Of Physiological Sciences. 2012;62:147–161. doi: 10.1007/s12576-012-0192-0.
Ohta T, Masuda K, Hara T, Tsujii J, Tsuruoka Y, Takeuchi J, Kim J.-D, Miyao Y, Yakushiji A, Yoshida K, Tateisi Y, Ninomiya T. An intelligent search engine and GUI-based efficient MEDLINE search tool based on deep syntactic parsing. In: Proceedings of the COLING/ACL on Interactive Presentation Sessions. Morristown, NJ, USA: Association for Computational Linguistics; 2006:17–20. doi: 10.3115/1225403.1225408.
Pang B, Lee L. Opinion mining and sentiment analysis. Foundations and Trends® in Information Retrieval. 2008;2:1–135. doi: 10.1561/1500000011.
Papazoglou M. Web Services: Principles and Technology 752. 2008.
Perez-Iratxeta C, Bork P, Andrade M.A. XplorMed: a tool for exploring MEDLINE abstracts. Trends in Biochemical Sciences. 2001;26:573–575.
Rebholz-Schuhmann D, Kirsch H, Arregui M, Gaudan S, Riethoven M, Stoehr P. EBIMed–text crunching to gather facts for proteins from Medline. Bioinformatics. 2007;23:e237–e244. doi: 10.1093/bioinformatics/btl302.
Reid E. US domestic extremist groups on the web: link and content analysis. IEEE Intelligent Systems. 2005;20:44–51. doi: 10.1109/MIS.2005.96.
Ridnour L.a, Dhanapal S, Hoos M, Wilson J, Lee J, Cheng R.Y.S, Brueggemann E.E, Hines H.B, Wilcock D.M, Vitek M.P, Wink D.a, Colton C.a. Nitric oxide-mediated regulation of β-amyloid clearance via alterations of MMP-9/TIMP-1. Journal of Neurochemistry. 2012;123:736–749. doi: 10.1111/jnc.12028.
Robillard R, Riopelle J.L, Adamkiewicz L, Tremblay G, Genest J. Pulmonary complications during treatment with hexamethonium. Canadian Medical Association Journal. 1955;72:448–451.
Russell D.G, VanderVen B.C, Lee W, Abramovitch R.B, Kim M, Homolka S, Niemann S, Rohde K.H. Mycobacterium tuberculosis wears what it eats. Cell Host Microbe. 2010;8:68–76. doi: 10.1016/j.chom.2010.06.002.
Rzhetsky A, Seringhaus M, Gerstein M. Seeking a new biology through text mining. Cell. 2008;134:9–13. doi: 10.1016/j.cell.2008.06.029.
Smalheiser N.R, Zhou W, Torvik V.I. Anne O’Tate: a tool to support user-driven summarization, drill-down and browsing of PubMed search results. Journal of Biomedical Discovery and Collaboration. 2008;3:2. doi: 10.1186/1747-5333-3-2.
Stableforth D.E. Chronic lung disease. Pulmonary fibrosis. British journal of hospital medicine. 1979;22(128):132–135.
Steinberger R, Fuart F, Goot E., Van Der, Best C. Text Mining from the Web for Medical Intelligence. San Fr: Heal; 2008 doi: 10.3233/978-1-58603-898-4-295 295–310.
Thuong N.T.T, Dunstan S.J, Chau T.T.H, Thorsson V, Simmons C.P, Quyen N.T.H, Thwaites G.E, Lan N.T.N, Hibberd M, Teo Y.Y, Seielstad M, Aderem A, Farrar J.J, Hawn T.R. Identification of tuberculosis susceptibility genes with human macrophage gene expression profiles. PLoS Pathogens. 2008;4 doi: 10.1371/journal.ppat.1000229.
Toda N. Regulation of blood pressure by nitroxidergic nerve. Journal Of Diabetes And Its Complications. 1995;9:200–202.
Van der Sar A.M, Spaink H.P, Zakrzewska A, Bitter W, Meijer A.H. Specificity of the zebrafish host transcriptome response to acute and chronic mycobacterial infection and the role of innate and adaptive immune components. Molecular Immunology. 2009;46:2317–2332. doi: 10.1016/j.molimm.2009.03.024.
Wilbur W.J, Hazard G.F, Divita G, Mork J.G, Aronson A.R, Browne A.C. Analysis of biomedical text for chemical names: a comparison of three methods. Proceedings of AMIA Symposium. 1999:176–180.
Wollmer M.A, Papassotiropoulos A, Streffer J.R, Grimaldi L.M.E, Kapaki E, Salani G, Paraskevas G.P, Maddalena A, de Quervain D, Bieber C, Umbricht D, Lemke U, Bosshardt S, Degonda N, Henke K, Hegi T, Jung H.H, Pasch T, Hock C, Nitsch R.M. Genetic polymorphisms and cerebrospinal fluid levels of tissue inhibitor of metalloproteinases 1 in sporadic Alzheimer’s disease. Psychiatric Genetics. 2002;12:155–160.
Yamamoto Y, Takagi T. Biomedical knowledge navigation by literature clustering. Journal of Biomedical Informatics. 2007;40:114–130. doi: 10.1016/j.jbi.2006.07.004.
Yan P, Hu X, Song H, Yin K, Bateman R.J, Cirrito J.R, Xiao Q, Hsu F.F, Turk J.W, Xu J, Hsu C.Y, Holtzman D.M, Lee J.-M. Matrix metalloproteinase-9 degrades amyloid-beta fibrils in vitro and compact plaques in situ. Journal of Biological Chemistry. 2006;281:24566–24574. doi: 10.1074/jbc.M602440200.
Yeasin M, Malempati H, Homayouni R, Sorower M. A systematic study on latent semantic analysis model parameters for mining biomedical literature. BMC Bioinformatics. 2009;10:A6. doi: 10.1186/1471-2105-10-S7-A6.
Yong V.W, Krekoski C.A, Forsyth P.A, Bell R, Edwards D.R. Matrix metalloproteinases and diseases of the CNS. Trends in Neuroscience. 1998;21:75–80. doi: 10.1016/S0166-2236(97)01169-7.
Zuberi K, Franz M, Rodriguez H, Montojo J, Lopes C.T, Bader G.D, Morris Q. GeneMANIA prediction server 2013 update. Nucleic Acids Research. 2013;41:W115–W122. doi: 10.1093/nar/gkt533.