Building research data handling systems with open source tools
Abstract:
Pharmaceutical discovery and development requires handling of complex and varied data across the process pipeline. This covers chemical structures information, biological assay and structure versus activity data, as well as logistics for compounds, plates and animals. An enterprise research data handling system must meet the needs of industrial scientists and the demands of a regulatory environment, and be available to external partners. Within Lundbeck, we have adopted a strategy focused on agile and rapid internal development using existing open source software toolkits. Our small development team developed and integrated these tools to achieve these objectives, producing a data management environment called the Life Science Project (LSP). In this chapter, I describe the challenges, rationale and methods used to develop LSP. A glimpse into the future is given as we prepare to release an updated version of LSP, LSP4All, to the research community as an open source project.
1.1 Introduction
All pharmaceutical company R&D groups have some kind of ‘corporate database’. This may not originally be an in-house designed knowledge base, but is still distinct from the specific area tools/databases that companies acquire from different software vendors. The corporate database is the storage area for all the ‘final’ pre-clinical results that companies want to retain indefinitely. The corporate database holds data from chemistry, biology, pharmacology and other relevant drug discovery disciplines and is also often a classic data warehouse [1] in the sense that no transactions are performed there – data is fed from the other databases, stored and retrieved. The system in this chapter is partly an example of such an infrastructure, but with a somewhat unique perspective.
The system is not only a data warehouse, however. Final/analysed data from other (specialist) tools are uploaded and stored there. Additionally, it is the main access point for data retrieval and decision support, but the system does a lot more. It forms the control centre and heart of our data transactions and workflow support through the drug discovery process at Lundbeck [2]. Lab equipment is connected, enabling controlled file transfer to equipment, progress monitoring and loading of output data directly back into the database. All Lundbeck Research logistics are also handled there, covering reagents, compounds, plates and animals. The system is updated when assets enter the various sites and when they get registered, and it stores location information and handles required re-ordering by scientists.
The system also supports our discovery project managers with ‘project grids’ containing compounds and assay results. These project grids or Structure Activity Relationship (SAR) tables are linked to the research projects and are where the project groups setup their screening cascade, or tests in which they are interested. Subsequently, the project groups can register both compounds and assay results to generate a combined project results overview. The grids also enable simple data mining and ordering of new tests when the teams have decided what compounds should be moved forward. To read more about corporate pharmaceutical research systems see references [3, 4].
How is the system unique? Is this any different from those of other companies? We believe it is. It is one coherent system built on top of one database. It covers a very broad area with data concerning genes, animals and compounds in one end of the process all the way to the late-stage non-GLP/GMP [5] exploratory toxicology studies. With a few exceptions, which are defined later, it is built entirely with open source software. It is therefore relevant to talk about, and fits well with the theme of this book, as a case in which a pharmaceutical company has built its main corporate database and transaction system on open source tools.
Corporate sales colleagues would likely call our system something like enterprise research data management or ‘SAP [6] for Research’. We simply call it LSP – which is short for the Life Science Project.
1.2 Legacy
It is difficult to make a clear distinction between before and after LSP, as the core part of the database was initiated more than 10 years ago. Internally, LSP is defined as the old corporate database combined with the new user interface (UI) (actually the full stack above the database) as well as new features, data types and processes/workflows. The following section describes what our environment looked like prior to LSP and what initiated the decision to build LSP.
Lundbeck has had a corporate database combining compounds and assay results since 1980. It has always been Lundbeck’s strategy to keep the final research data together in our own in-house designed database to facilitate fast changes to the system if needed, independently of vendors.
Previously, research used several closed source ‘speciality’ software packages with which the scientists interacted. In chemistry these were mainly centred around the ‘ISIS suite’ of applications from what used to be called MDL [7]. They have since been merged into Symyx [8], which recently became Accelrys [9]. As an aside, this shows the instability of the chemistry software arena, making the decision to keep (at least a core piece of) the environment in-house developed and/or in another way independent of the vendors more relevant. If not in-house controlled/ developed, then at least using an open source package will enable a smoother switch of vendor if the initial vendor decides to change direction.
The main third-party software package in the (in vitro) pharmacology area was ActivityBase [10], a very popular system to support plate-based assays in pharma in the early 2000s. Whereas the ISIS applications were connected to the internal corporate database, ActivityBase came with its own Oracle database. Therefore, when the chemists registered compounds into our database the information about the compounds had to be copied (and hence duplicated) into the ActivityBase database to enable the correct link between compounds and results. After analysis in ActivityBase, the (main) results were copied back into our corporate database. Hardly efficient and lean data management!
Of course, the vendors wanted to change this – by selling more of their software and delivering the ‘full enterprise coverage’. Sadly, their tools were not originally designed to cover all areas and therefore did not come across as a fully integrated system – rather they were a patchwork of individual tools knitted together. A decision to move to a full vendor system would have been against Lundbeck’s strategy, and, as our group implemented more and more functionality in the internal systems, the opposite strategy of using only in-house tools became the natural direction.
Workflow support is evidently a need in drug discovery. Scientists need to be able to see the upstream data in order to do their work. Therefore, ‘integration projects’ between different tools almost always follow after acquisition of an ‘of the shelf’ software package. The times where one takes software from the shelf, installs and runs it are truly rare. Even between applications from the same vendor – where one would expect smooth interfaces – integration projects were needed.
As commercial tools are generally closed source, the amount of integration work Lundbeck is able to do, either in-house or through hired local programmers with relevant technology knowledge is very limited. This means that on top of paying fairly expensive software licences, the organisation has to hire the vendor’s consultants to do all the integration work and they can cost £1000/day. If one part of the workflow is later upgraded, all integrations have to be upgraded/re-done resulting in even more expensive integration projects. Supporting such a system becomes a never-ending story of upgrading and integrating, leaving less time for other optimisations.
At that time the internal Research Informatics UI offering was a mixture of old Oracle Forms-based query and update forms and some Perl web applications that were created during a period of trying to move away from Oracle Forms. However, this move towards fully web-based interfaces was put on hold when Lundbeck decided to acquire Synaptic Inc. [11] in 2003, leading to prioritisation of several global alignment and integration projects into the portfolio.
Clearly, this created an even more heterogeneous environment, and an overall strategic and architectural decision, enabling us to build a coherent affordable and maintainable system for the future, was needed.
1.3 Ambition
With the current speed of technology developments, to think that one can make a technology choice today and be ‘prepared for the future’ is an illusion. Under these circumstances, how does one choose the right technology stack?
Very few have the luxury of basing a technology decision purely on objective facts. Firstly, it takes a long time to collect these facts about the possible options (and in some cases the objective truth can only be found after coding the application in several technologies and comparing the results, and this is very rarely done). Secondly, it is a fact that a technology decision is always taken in a certain context or from a certain point of view, making these kinds of decisions inherently biased. Finally, according to research described in the book Sway [12] we all, in general, overrate our rationality in our decision making.
Therefore, any decision is likely to be based on a combination of objective facts, current environment, current skill sets in the team, what the infrastructure strategy/policy allows and whether we like it or not; our gut feeling! Despite our best efforts and intentions, in the end the right choice becomes clear only after a couple of years, when it is apparent which technologies are still around, have been adopted by a significant number of groups and are still in active development.
With the above decision limitations in mind, the following are the criteria used when deciding on technology for the future global Lundbeck research system.
![]() Multi-tier architecture – an architecture where the client, running on local PCs, is separated from the application server, which is separated from the database server. In our case, the servers should be located only at Lundbeck HQ in Denmark, keeping the hardware setup as simple and maintainable as possible.
Multi-tier architecture – an architecture where the client, running on local PCs, is separated from the application server, which is separated from the database server. In our case, the servers should be located only at Lundbeck HQ in Denmark, keeping the hardware setup as simple and maintainable as possible.
![]() Web-based UI – a web-based solution that would be easy to deploy on all the different kinds of PCs and languages used around the world. A lightweight web client should still be fast enough from any location with the servers placed in Denmark.
Web-based UI – a web-based solution that would be easy to deploy on all the different kinds of PCs and languages used around the world. A lightweight web client should still be fast enough from any location with the servers placed in Denmark.
![]() Open source – open source tools give us the confidence that we are likely always to find a solution to the problems we encounter, either by adding a plug-in if one is available or by developing what is needed in-house.
Open source – open source tools give us the confidence that we are likely always to find a solution to the problems we encounter, either by adding a plug-in if one is available or by developing what is needed in-house.
![]() Oracle – as the existing data are stored in a relational database based on Oracle, we need a technology with support for this type of database.
Oracle – as the existing data are stored in a relational database based on Oracle, we need a technology with support for this type of database.
In addition, there are also more subjective, but no less important, criteria to consider.
![]() Fast to develop – a web framework that is fast to learn/get started with and fast when developing, relative to other technology choices.
Fast to develop – a web framework that is fast to learn/get started with and fast when developing, relative to other technology choices.
![]() Active development – tools having a large and active development community.
Active development – tools having a large and active development community.
![]() Easy to deploy – with only limited requirements on the user platform in terms of Operating System, browser etc., hence reducing the probability and impact of desktop platform upgrades.
Easy to deploy – with only limited requirements on the user platform in terms of Operating System, browser etc., hence reducing the probability and impact of desktop platform upgrades.
In conclusion our ambition was to build a web-based research enterprise data management system, covering the pre-clinical areas at Lundbeck, entirely on open source tools with the exceptions that the legacy commercial database and chemical engine would stay.
1.4 Path chosen
One could argue that an in-house system such as LSP is commodity software that does not offer anything more than some commercial packages, and therefore should be bought ‘off the shelf’. It is argued here that there was (is) no single commercial system available which supports all the areas of functionality required across the pipeline described in the introduction. Consequently, without our own in-house system Lundbeck would need to acquire several different packages and subsequently combine these via continued expensive integration projects as mentioned earlier. Such an approach is very likely not the best and most cost-effective approach and certainly does not give the scientists the best possible support. Additionally, the maintenance of many single applications would be a burden for the department. Thus, the approach at Lundbeck is instead to build one coherent system. Everything that is built needs to fit into the system and hence all projects are extensions of the system rather than individual applications for chemistry, biology and pharmacology.
Having a one-system focus is not without conflicts in today’s business environment with a heavy focus on projects, deliverables and timelines (not to mention bonuses). If developers are forced to deliver on time and on budget they will likely ask for agreed deliverables, which means a small(er) tangible piece of code (application) on which all stakeholders can agree. Firstly, if we can agree up front what needs to be built we are not talking about something new (hence no innovation), and, secondly, if the timeline and/or budget is approaching the developers will cut down on the functionality. The question is whether software developed under these circumstances will match the real requirements.
The one-system approach allows searches across data from different areas without any integration, data warehousing or data duplication. We can maintain the environment with a small group of people as maintenance and development becomes ‘one and the same’. There is only a need for one group – not a group to develop and another to maintain. Since 2004, the Research Informatics group has consisted of seven people (six developers and the head) placed at two sites supporting around 400 researchers, as well as data input from five to ten CRO partners around the world.
The aim is to get more coding and support for less money and people. The Informatics group is placed in the centre of a network from where we import and utilise the relevant tools from the network for incorporation into our LSP platform. This setup is a combination of ‘re-use and build’ as very little is coded from scratch. Instead, we use libraries, tools and frameworks that others develop and maintain while we concentrate on adding domain expertise and the one-system vision to combine all tools in the right way for us.
1.5 The ‘ilities
In system engineering (software development) two different types of requirements exist – the functional and the non-functional. The nonfunctional, as the name indicates, does not concern what the system needs to do for the user but rather how it does it. There are many more or less important non-functional requirements like scalability, availability and security often collectively called ‘the ‘ilities’ [13].
Interestingly there is a tendency for management, IT, consultants etc., to put the ‘ilities forward when talking about outsourcing, cloud computing, open source software or anything else new that might disrupt the ‘as is’ situation and shift people out of the comfort zone or potentially cause a vendor to lose business. This is a well-known sales strategy described as FUD (Fear, Uncertainty and Doubt) marketing.
The part of the non-functional requirements that are viewed as important by an organisation must apply equally to open as well as closed source software. But this does not always seem to be the case. Often the open source tools may receive more scrutiny than the closed source commercial ones. It is not a given that a commercial closed source tool is more scalable, reliable or secure than other pieces of software. Certainly poor open source tools are available, as are good ones – just as there are poor commercial tools and good commercial tools.
There is a tendency in the pharmaceutical industry to aim too high in terms of the non-functional requirements. Naturally, the tools deployed need to be scalable and fast. But one must be sure to add the relevant amount of power in the relevant places. As an example, what happens if the corporate database is unavailable for a couple of days? Scientists can continue their lab experiments, can read or write papers and prepare their next presentation. The research organisation does not come to a complete stop. Is 99.99% availability really needed and does it make sense to pay the premium to get the last very expensive parts covered? The experience of our group suggests otherwise, we assert that many of the relevant open source tools are sufficiently stable and scalable to meet the needs of pharmaceutical research.
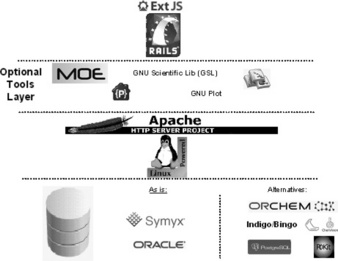
After these considerations – and some initial reading about the different tools, to understand how they really worked, and try to get a feel for the level of activities in the different communities – it was decided to go with the stack outlined in Figure 1.1 and described in detail below.
1.5.1 Database
When deciding on the LSP stack, a database change was ruled out. The Oracle database is one of the exceptions from the fully open source stack mentioned in the introduction. It would be natural to build LSP on top of an open source database but at the time a deliberate decision was made to continue our use of Oracle. As the Lundbeck corporate IT standard database, we are able to utilise full support and maintenance services from IT, not to mention the many years of experience and existing code we have developed. However, the main reason for not moving to an open source database was that this would trigger a switch of the other non-open source piece, the chemistry engine from Symyx/Accelrys, which did not support any of the open source databases. Changing a chemistry engine requires a lot of testing to make sure the functionality is correct and also a lot of change management related to the scientists who need to change their way of working. We did not have the resources to also handle a chemistry engine change on top of all the other technology changes and hence decided to focus on the remaining technologies.
1.5.2 Operating system (OS)
Based on our internal programming expertise, it was natural to choose Linux [14] as the OS for the web server component. It ensures the ability to add the functionalities/packages needed and Linux has proven its value in numerous powerful installations. Linux is used in 459 of the top 500 supercomputers [15] and as part of the well-known LAMP-stack [16].
In terms of Linux distributions the department’s favourites are Debian [17] based. However, it was decided to use the one delivered by our Corporate IT group, RedHat Enterprise Linux [18], as part of their standard Linux server offerings. Service contracts for RedHat were already in place and we could still install the tools we needed. Corporate IT currently run our databases and servers, handle upgrades and backups while we concentrate on adding tools and perform the programming.
1.5.3 Web server
The choice of web server might have been even easier than choosing the OS. Firstly, Apache [19] has for several years been the most used web server in the world and is, according to the Netcraft web server survey [20] July 2011, hosting 235 million web sites. Secondly we already had experience with Apache as we used it to support our Perl environment.
1.5.4 Programming language
We did not choose Ruby [21] as such; we chose the web framework (see next section), which is based on the Ruby language. Before deciding to use the web framework and hence Ruby, we looked quite carefully into the language. What do people say about it? What is the syntax? Does it somehow limit our abilities especially compared to C [22], which we earlier used to perform asynchronous monitoring of lab equipment and similar low-level TCP/IP communication. Our initial investigations did not lead to any concerns, not even from the experienced colleagues trained in many of the traditional languages.
Ruby may have been on the hype curve [23] at this time but it is not a new language. It has been under careful development over more than 15 years by Matz [24], with the aim of combining the best from several existing languages.
1.5.5 Web framework
Several years of Perl programming had resulted in a lot of code duplication. The new web framework needed to be an improvement from our Perl days, providing clear ideas for separation and re-use rather than copy code.
The Ruby on Rails (RoR) (http://rubyonrails.org/) framework had been getting a lot of attention at the time, with a focus on the following concepts.
![]() Convention over configuration.
Convention over configuration.
![]() REST – or Representational State Transfer, which is the basis for a RESTful architecture [25], seen by many as the best way to implement web applications today.
REST – or Representational State Transfer, which is the basis for a RESTful architecture [25], seen by many as the best way to implement web applications today.
Ruby on Rails was initiated by David Heineman Hansson (DHH) [26] as part of the Basecamp [27] project. In general, the framework looked very productive and the vision/thinking from the people behind RoR/37Signals about software development fit very well with our internal thinking and way of working. As it appeared easy to use and there seemed to be high adoption and very active development, we decided that the future research informatics development at Lundbeck should be based on RoR.
Although one may download RoR freely, the choices of how to serve the graphical interface, organise modules and workflows, pass data between modules and many other considerations are open and complex. Thus, our web framework is more than just RoR, it is a Ruby-based infrastructure for defining, connecting and deploying distinct data service components.
1.5.6 Front-end
UI design is inherently difficult. It takes a lot of time and is rarely done well. One can appreciate good design but it is not as easy to create. The tools need to look good or ‘professional’, otherwise the users will judge the functionality harshly. A nice looking front-end is required in addition to correct functionality. RoR comes with a clean but also very simple UI, but an interface with more interactivity was required (implemented using AJAX/javascript) to enable the users to work with the data.
The javascript library Extjs [28] delivers a rich UI framework with pre- build web tools such as grids, forms and tables that can be used to build the special views needed for the different data types. Extjs also creates a platform on which many external javascript tools such as structure viewers can be incorporated via a ‘plug-in’ mechanism. Although Extjs is an open source library, it is not free for all due to licence restrictions on commercial use. However, on open source projects, developers can use Extjs for free under a GPL [29] type licence [30].
1.5.7 Plug-ins
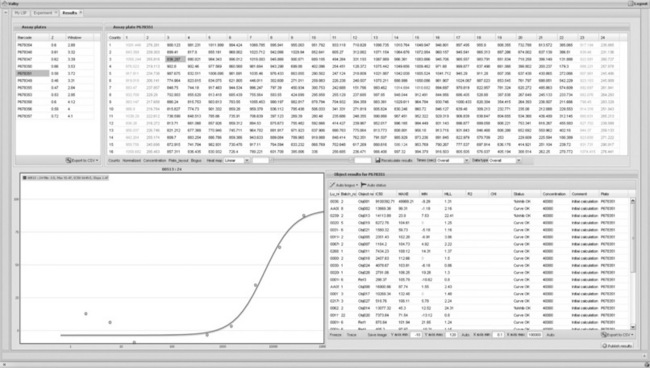
LSP also utilises different plug-ins for some special tasks. As an example, the GNU Scientific Library (GSL) [31] is used as support for simple maths and statistics calculations. Many of the standard calculations performed on science data are fairly straightforward and well described, and can be implemented with an open source math library and some internal brainpower. With these tools, IC50-type sigmoidal curve fitting has been implemented using the flot [32] javascript library for graphics rendering in the browser allowing scientists to see and interact with the data (Figure 1.2).
1.5.8 Github
All the LSP code is hosted on Github [33], a low-cost (free for open source projects) and effective site hosting software projects using the Git source code version control system [34]. Git was, like Linux, initially developed by Linus Torvalds [35]. It is fully distributed and very fast, and many of the tools selected made use of Git, and were already hosted on Github. It would therefore be easier to merge future, new code and technologies into the code stream by also using Git. Despite the fact that it added another complexity and another new thing to learn, it was decided to follow the RoR community and move to Git/Github.
As mentioned, most, but at the moment not all, parts of the stack are open source tools. An Oracle database is used, whereas structure depictions and searches use tools from Symyx/Accelrys. MOE [36] is also not a FLOSS tool. At Lundbeck it is used for some speciality compound calculations where the result is displayed in LSP. MOE is not essential for the running of LSP, but Oracle and the Symyx/Accelrys cartridge (for chemical structures) currently are.
1.6 Overall vision
LSP is designed to span the entire continuum, from data storage and organisation, through web/query services and into GUI ‘widgets’ with which users interact. The core of the system is a highly configurable application ‘frame’, which allows different interface components (driven by underlying database queries) to be connected together and presented to the user as business-relevant forms. Meta-data concerning these components (which we call ‘application modules’) are stored in the LSP database, enabling us to re-use and interlink different modules at will. For example, connecting a chemical structure sketcher, pharmacology data browser and graph widget together for a new view of the data. It is this underlying framework that provides the means to develop new functionality rapidly, with so much of the ‘plumbing’ already taken care of. With this in place, our developers are free to focus on the more important elements, such as configuring the data storage, writing the queries and deploying them in user-friendly interface components.
1.7 Lessons learned
The informatics department consists of experienced programmers, who tend to learn new tools, languages etc., proactively by reading an advanced-level book and starting to work with the new skills, i.e. learning by doing. RoR was no exception. The team acquired some of the books written by DHH and started the development. It is evident, as can also be seen on the RoR getting started page [37] that RoR is a really quick and easy way to build something relatively simple.
In order to achieve the intended simplicity, RoR expects simple database tables following a certain naming scheme – if the model is called Compound the table behind should be Compounds in plural. This is convention over configuration. If one already has a big database in place that does not follow this naming scheme, some of the ease of use and smartness of RoR disappears. This was true in our case. Tough architecture discussions therefore started when choosing between changing the database to fit RoR and adapting RoR to our existing setup. The database was the only remaining piece of the old setup, so it was desirable that this should not enforce the mindset of the old world while using technologies from the new world, which might have happened in this case.
Building LSP is more than just downloading and installing the key components. They have to be built together in a meaningful way in which the different parts handle the tasks they are designed to do, and do best, in the relevant context. But it is not always clear where to make a clean separation of the layers to achieve the best possible solution. In addition, programmers may often disagree. RoR follows the Model-View-Controller [38] paradigm. In the RoR world this, simply explained, means a model that gets the data from the database, a view that takes care of the design of the page displayed to the user and a controller that glues the pieces together. Although it would have been preferable to follow that layout, it was again realised that our mature database would force some tough architecture decisions. The database contains many views as well as Oracle PL/SQL procedures. Should the procedures be replaced with models and controllers? Would it be better to have a simple database and then move all sorts of logic into the RoR layer? After some consideration, the real world answer is that ‘it depends’!
Oracle procedures are good for transactions inside the database. Taking data out of the database, manipulating them in the RoR layer and putting them back into the database did not seem a very efficient approach. On the other hand, it is preferable not to be single-database-vendor centric. Moving to a simpler table/procedure structure would likely make it easier to port to other databases if/when that becomes relevant.
Similarly, how to best combine RoR and Extjs was debated. As mentioned, RoR delivers a way to build interfaces but using Extjs also meant finding a way to elegantly knit the two layers together. A decision needed to be taken regarding incorporating the Extjs code into the view part of the RoR MVC setup or keeping it separate. Integrating Extjs into RoR would tie the two parts very closely together and although that might have advantages, it would make it more complicated to replace one of the layers if something better were to appear at a later stage. While deciding how to integrate Extjs with RoR, we also needed to learn how to use the Extjs library itself most effectively.
It took time to agree/understand how to best use and integrate the layers of the stack and we did not get to that level of understanding without a longer period of actual coding. The current LSP code base may not currently be structured in the best possible way for long-term support and development, similar code structure issues are known and understood by other developers in the community. The future direction of RoR and Extjs are consistent with the Lundbeck team’s views of the ‘right way’ to structure and enhance the code in future. The team behind Extjs recognised that they needed to implement more structure in their library in order to support the various developers. A MVC structure is therefore implemented in version 4 of Extjs. Hence a clean installation of the new version of our stack is being considered, along with re-building LSP as that will be easier, better and/or faster than trying to upgrade and adapt our initial attempt to fit the new version of the stack.
1.8 Implementation
It is likely that there is never going to be a ‘final’ implementation. The development is going to continue – hence the name Life Science Project – as technologies and abilities change as do the needs in terms of more data types, analysis of the data, better ways of displaying the data, etc.
Overall, the current system looks very promising. Our group is still happy with the choices made regarding the tools to use and LSP does work very well. It is therefore entirely possible, and in fact very beneficial and cost-effective, to develop a research enterprise data management system purely on open source tools. All parts of the technology stack continue to develop, and Ruby, and especially RoR and Extjs, are moving forward at high speed and adding a lot of new valuable features.
Neither scale nor speed or any other of the ‘ilities seem to be an issue with our implementation and number of users. Occasional slowness in the front-end is due to (1) very big/complex database queries that need optimisation or can be handled with more power on the database or (2) Javascript rendering on the clients. This is not an issue in general but certain (older) PCs with lack of RAM and/or certain browsers do not render quickly enough. On a standard laptop running Windows and using Firefox, LSP works very well.
Using Javascript (a Javascript library) as the UI has also turned out to be a very sound idea. Javascript can be challenging to work with, but, handled correctly, it can be used to create many relevant tools [39]. There are a lot of useful javascript-based visualisations and plug-ins on the web that are easy to incorporate into LSP, making the front-end attractive as well as interactive with several different ways to display the relevant data to the scientists.
1.9 Who uses LSP today?
Years of experience have shown that creating something new for users, in a new technology that is only slightly better than what they currently use is not the road to success. Users do not seem to think the new technology is as exciting as do the development team! For the LSP deployment it was decided to only involve the users when LSP would give them something new or greatly improve what they already had.
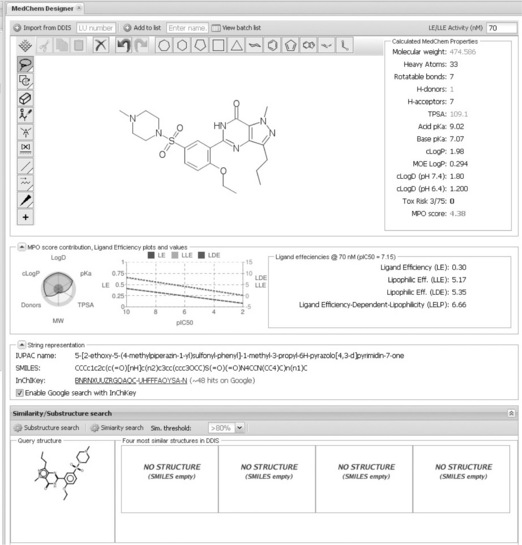
The first piece deployed to a larger group was our interactive (real time) MedChem Designer (Figure 1.3). The medicinal chemists responsible for designing pre-clinical compounds are able to draw structures that the system will submit to a property calculator and return the results in easy readable visualisations. The response time is instant, meaning as soon as the chemists change the structure in any way the properties will be updated. This was a tool Lundbeck did not previously have, it was liked by chemists, leading to almost immediate and full adoption in the chemistry groups.
The informatics group handles all logistics in research (compounds, plates, animals, etc.) but the chemistry reagent logistics were still being handled by an old ISIS-based application that was performing poorly. The existing in-house compound logistics database functionality was re-used and a new front-end with up-to-date functionality was created in LSP, also incorporating a new workflow that would support reagent ordering by local chemists but shipment directly to outsourcing partners. Because of the huge increase in usability and functionality, and by migrating systems over a weekend, an easy deployment with full adoption was achieved.
One benefit of investing time in re-building a fairly small reagent application was also that it would enable a move of compound/reagent registration into LSP. This was not an expressed need from the chemists but was required to enable direct registration of compounds from our outsourcing partners into the LSP database in Denmark. Shortly after the reagent store deployment, the LSP compound registration form was published as a Lundbeck extranet solution, where the chemistry partners can access and use it directly (Figure 1.4). This is now deployed to five different partner companies placed in three different countries on several continents around the world and works very well. This platform therefore enables Lundbeck to very quickly and cost-effectively implement direct CRO data input, something basically all pharmaceutical companies need today. The LSP partner solution, LSP4Externals, is a scaled-down (simpler) version of the internal solution. Security considerations resulted in the deployment of a separate instance, although using the same database. A row level security implementation makes it possible to give the different partners access to subsets of compounds and/or assays in the projects as relevant.
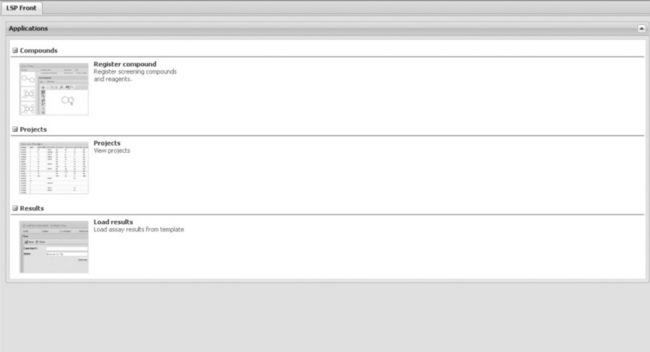
Figure 1.4 LSP4Externals front page with access to the different functionalities published to the external collaborators
Although assay registration was implemented fairly early in LSP, it was not deployed as a general tool. Assay registration functionality existed in the old systems, although it was not as effective and the new LSP version therefore did not initially give the users significant new functionality. The LSP version of assay registration would introduce a major, but necessary data cleanup. Unfortunately, an assay information cleanup introduces more work, not functionality, to the scientists, so ‘an award’ was needed that would show the user benefits of this exercise. The award was a new assay query tool that enabled searches across parameters from all global research assays and therefore made it easier for the scientists to find all assays working against a certain target, species, etc.
This started the move towards using LSP for assay registration and retrieval. Another important push came with external (partner) upload of data and collaboration on project grids/SAR tables. In order to make sure the external partners can understand and use the Lundbeck data, the scientists need to clean up the assay information.
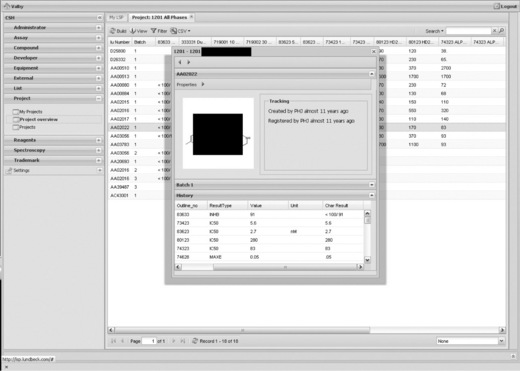
The last part of existing legacy functionality to be moved into LSP and then improved, is our project overview/SAR table grids, displaying the combined view of compounds and results in our projects. This should be the centre/starting point for all data retrieval and analysis as well as transactions in the form of new assay or compound ordering. Currently, the project overview with the ability to see all history data on a single compound has been ported to LSP (Figure 1.5), while ordering is in the plans.
1.10 Organisation
IT/Informatics departments normally consist of managers, business analysts, project managers, applications specialists, etc. as well as developers (if still available in-house). The departments therefore tend to become large and costly while most of the individuals do not develop or maintain applications but rather manage resources. However, the biggest issue with these types of organisations are the translations, where valuable information gets lost. There are translations from scientists to business analysts, from analysts to project managers, from project managers to developers – or more likely from project managers to the outsourcing partner project managers and then to the developers. The handovers between all these individuals are normally performed via a User Requirement Specification (URS). The developer therefore has to build what is described in the document instead of talking directly to the users and seeing them in action in the environment in which the system is going to be used.
Lundbeck Research Informatics uses another approach, without any business analysts or any other type of translators and without any URS documents. All members of the team are code-producing and in order for them to understand what is needed they will visit the relevant scientists, see them in action and learn their workflow. This way the programmers can, with their own eyes, see what the scientists need to do, what data they require and what output they create. When asked to identify their requirements, users may reply ‘what we have now, only bigger and faster’. Users naturally do not see a completely new solution they did not know existed, when they are deep into their current way of doing things. Do not build what they want, but what they need. It is evident that users are not always right!
This way of working naturally requires skilled people with domain knowledge as well as programming skills. They all constantly need to monitor the external communities for new relevant tools, incorporate the tools into the environment in a meaningful way and build systems that make sense to scientists. We therefore try to combine different types of ‘T-shaped’ [40] people. They are either experienced programmers with science understanding or scientists with programming understanding.
Our group operate as a small, close-knit team, ensuring that ‘everybody knows everything’ and therefore avoid any unnecessary alignment meetings. Meetings are normally held in front of a white board with the entire team present and concern workflow or front-end design, so all members know what is going on, what is being decided and everybody can contribute ideas.
The method is not unique and was not invented by us, although it may be unique in the pharma world, in Lundbeck Research Informatics it is well established and feels natural. Similar approaches are being used by several successful companies within innovation, design and software development like IDEO [41] and 37signals [42]. In many of the large open source projects that require some form of coordination, all participants are also still producing and/or at least reading and accepting the code entering the code base.
In addition, the mode of action is dependent on empowerment and freedom to operate. If the informatics group needed to adhere to corporate technology policies or await review and acceptance before embracing a new tool, it would not be as successful. At Lundbeck the group reports to the Head of Research and hence is independent of Corporate IT and Finance as well as the individual disciplines within Research.
The group has been fortunate to experience only a single merger and therefore has had the opportunity to focus on the one-system strategy for a longer period, with the environment and system kept simple enough for us to both develop and maintain at the same time.
1.11 Future aspirations
We do not expect, and do not hope, that the Life Science Project will ever end. As long as Lundbeck is actively engaged in research there will be a constant need for new developments/upgrades with support for new data types, new visualisations and implementation of new technologies that we do not yet know about.
Currently, we are looking into the use of semantic technology [43]. What is it and where does it make most sense to apply? Linking our internal databases via Resource Description Framework (RDF) might enable us to perform searches and find links in data that are not easily found with the current SQL-based technologies. But, as mentioned here, a lot of our research data are stored together in one database so for that piece RDF might not be appropriate at the moment. The large amounts of publicly available external data that scientists use in their daily work, on the other hand, are good candidates for linking via RDF [44]. In order to continue to follow the one-system strategy, we are looking to display these linked external data sources in LSP alongside the internal data so the scientists can freely link to the external world or our in-house data.
With the new cloud-based possibilities for large-scale data storage and number crunching [45], a very likely future improvement to LSP will be use of a cloud-based setup for property calculations. In such a setup the scientists would submit the job and LSP would load the work to the cloud in an asynchronous way and return when the cloud calculator is done. The first cloud-based pilot projects have been initiated to better understand what the best future setup would look like.
Another new area to move into could be high-level management overview data. As all information is in one place, it would not be difficult to add more management-type views on the data. If overall research/ discovery projects have meta-data like indication, risk, phase added, we could then deliver portfolio-like overviews, which management currently produce by hand in Powerpoint. Chapter 17 by Harland and colleagues is relevant here, using semantic MediaWiki in managing disease and target knowledge and providing an overview to management. We also know how many compounds have been produced for the individual projects in what time period, by whom working for which partner and located at which site. This could give the project teams and management quick access to data about turnaround time and other efficiency metrics at the same front-end at which they can dive into raw data.
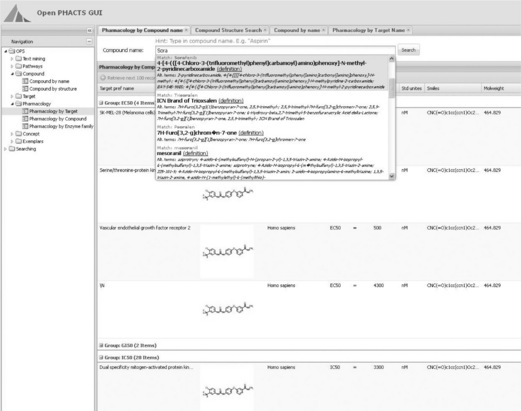
Lundbeck Research Informatics is participating in the Innovative Medicines Initiative (IMI) OpenPhacts project [46]. Our main contribution to the project is likely to be the front-end, which basically will be a scaled-down version of LSP, which we can further develop to fit the needs of the IMI project. The data in the project will be based on RDF and a triple store (Figure 1.6), so we will hopefully be able to improve the way LSP handles this type of data by means of the project and use that experience for our internal RDF data handling.
As true believers in open source software development, we feel obliged to give something back to the community. We therefore plan to open source the LSP platform development, under the name LSP4All, so others can download it and use it for data management in their labs, but naturally also because we thereby hope that others will join the team and help develop LSP further to the benefit of all future LSP users.
To see wide adoption and combined future development would be truly great. In order to encourage as wide an adoption of LSP as possible, we will make the LSP4All port based solely on FLOSS tools. That means moving away from the Oracle database and the Symyx/Accelrys chemical structure tools as we think these licence costs would deter many groups from using LSP4All. The move to LSP4All, or its initiation, has already started as we decided that porting LSP to LSP4All would be a great opportunity to create the scaled-down version of LSP needed to deliver to the OpenPhacts project, while at the same time re-building LSP using the new versions of our technologies in the preferred way for our internal use and also start an open source project. This was basically the window of opportunity we were looking for to start the LSP upgrade project.
Later, when sufficient LSP functionality has been moved, we expect LSP4All to be the version that will run here at Lundbeck. It follows our chosen design principles and it will save us some extra licence costs as it will be based fully on open source tools. The database currently in use for LSP4All is Postgresql [47], an open source database being used by many big corporations in many different industries [48] and therefore likely to support our database needs. Another, perhaps more natural, option would be MySQL [49]; however, following the acquisition of MySQL by Oracle there seems to be a tendency in the open source community to move to Postgresql.
The chemical structure needs of LSP4All will be handled by something like Ketcher [50], a pure open source Javascript-based editor, for structure depiction and like RDKit [51] for the structure searches. RDKit supports Postgrsql and therefore fits with the choice of open source database. Several new well-functioning open source chemistry packages and cartridges have appeared lately, so in the future there will be more options from which to choose.
The LSP4All project is hosted at Github and we expect to open the repository to the public during 2012. Then all university labs, small biotechs and other relevant organisations will have the opportunity to download and run a full data management package with chemistry support for free. It will be a big step towards more open source collaboration in the biopharmaceutical informatics space.
1.12 References
[1] Wikipedia Data warehouse. http://en.wikipedia.org/wiki/Data_warehouse. Updated 16 August 2011. Accessed 18 August 2011.
[2] Homepage of H. Lundbeck A/S. http://www.lundbeck.com. Accessed 18 August 2011.
[3] Agrafiotis, D.K., et al. Advanced Biological and Chemical Discovery (ABCD): centralizing discovery knowledge in an inherently decentralized world. Journal of Chemical Information and Modeling. 2007; 47:1999–2014.
[4] Pfizer Rgate – http://www.slideshare.net/bengardner135/stratergies-for-the-intergration-of-information-ipiconfex. Accessed 28 September 2011.
[5] Homepage of FDA – GLP Regulations. http://www.fda.gov/ICECI/EnforcementActions/BioresearchMonitoring/NonclinicalLaboratoriesInspectedunderGoodLaboratoryPractices/ucm072706.htm. Updated 8 May 2009. Accessed 28 September 2011.
[6] Homepage of SAP. http://www.sap.com/uk/index.epx#. Accessed 18 August 2011.
[7] Wikipedia MDL Information Systems. http://en.wikipedia.org/wiki/MDL_Information_Systems. Updated 3 December 2010. Accessed 18 August 2011.
[8] Wikipedia Symyx Technologies. http://en.wikipedia.org/wiki/Symyx_Technologies. Updated 2 December 2010. Accessed 18 August 2011.
[9] Homepage of Accelrys. http://accelrys.com/. Accessed 18 August 2011.
[10] Homepage of IDBS. http://www.idbs.com/activitybase/. Accessed 30 August 2011.
[11] Press release from H. Lundbeck A/S. http://lundbeck.com/investor/releases/ReleaseDetails/Release_89_EN.asp. Accessed 30 August 2011.
[12] Brafman, O., Brafman, R. Sway: The Irresistible Pull of Irrational Behavior, 1st ed. Crown Business; 2008.
[13] Wikipedia List of system quality attributes. http://en.wikipedia.org/wiki/List_of_system_quality_attributes. Updated 31 July 2011. Accessed 30 August 2011.
[14] The Linux Foundation. www.linuxfoundation.org. Accessed 30 August 2011.
[15] TOP500 Super Computer Sites. http://www.top500.org/stats/list/36/osfam. Accessed 30 August 2011.
[16] LAMP: The Open Source Web Platform. http://onlamp.com/pub/a/onlamp/2001/01/25/lamp.html. Updated 26 January 2001. Accessed 30 August 2011.
[17] Homepage of Debian. www.debian.org. Updated 30 August 2011. Accessed 30 August 2011.
[18] Homepage of RedHat Enterprise Linux. http://www.redhat.com/rhel/. Accessed 30 August 2011.
[19] Apache http server project. http://httpd.apache.org/. Accessed 30 August 2011.
[20] Web server survey. http://news.netcraft.com/archives/category/web-server-survey/. Accessed 18 July 2011.
[21] Ruby, A programmer’s best friend. http://www.ruby-lang.org/en/. Accessed 30 August 2011.
[22] Kernighan, B.W., Ritchie, D.M. C Programming Language, 2nd ed. Prentice Hall; 1988.
[23] Gartner Hype Cycle. http://www.gartner.com/technology/research/methodologies/hype-cycle.jsp. Accessed 30 August 2011.
[24] About Ruby. http://www.ruby-lang.org/en/about/. Accessed 30 August 2011.
[25] Architectural Styles and the Design of Network-based Software architectures. http://www.ics.uci.edu/fielding/pubs/dissertation/top.htm. Accessed 30 August 2011.
[26] About 37Signals. http://37signals.com/about. Accessed 30 August 2011.
[27] Homepage of Basecamp. http://basecamphq.com/. Accessed 30 August 2011.
[28] Sencha Extjs product site. http://www.sencha.com/products/extjs/. Accessed 30 August 2011.
[29] GNU General Public License. http://www.gnu.org/copyleft/gpl.html. Accessed 30 August 2011.
[30] Sencha Extjs Licensing Options. http://www.sencha.com/products/extjs/license/. Accessed 30 August 2011.
[31] GSL – GNU Scientific Library. http://www.gnu.org/software/gsl/. Updated 17 June 2011. Accessed 30 August 2011.
[32] Homepage of flot. http://code.google.com/p/flot/. Accessed 30 August 2011.
[33] Homepage of GitHub. www.github.com. Accessed 30 August 2011.
[34] Homepage of Git. http://git-scm.com. Accessed 30 August 2011.
[35] Torvalds, L., Diamond, D. Just for Fun: The Story of an Accidental Revolutionary, 1st ed. HarperCollins; 2001.
[36] Chemical computing group MOE product page. http://www.chemcomp.com/software.htm. Accessed 30 August 2011.
[37] Getting Started with Rails. http://guides.rubyonrails.org/getting_started.html. Accessed 30 August 2011.
[38] Applications Programming in Smalltalk-80™: How to use Model-View-Controller (MVC) http://st-www.cs.illinois.edu/users/smarch/st-docs/mvc.html – Updated 4 March 1997. Accessed 30 August 2011.
[39] JavaScript: The World’s Most Misunderstood Programming Language. http://www.crockford.com/javascript/javascript.html. Accessed 30 August 2011.
[40] IDEO CEO Tim Brown: T-Shaped Stars: The Backbone of IDEO’s Collaborative Culture. http://chiefexecutive.net/ideo-ceo-tim-brown-t-shaped-stars-the-backbone-of-ideoae%E2%84%A2s-collaborative-culture. Accessed 30 August 2011.
[41] Kelley, T., Littman, J. The Art of Innovation:Lessons in Creativity from IDEO, America’s Leading Design Firm. Crown Business; 2001.
[42] Fried, J., Hansson, D.H. Rework. Crown Publishing Group; 2010.
[43] Bizer, C., Heath, T., Berners-Lee, T., Linked Data – The Story So Far. International Journal on Semantic Web and Information Systems (IJSWIS). 2009;5(3), doi: 10.4018/jswis.2009081901. [accessed 13 September 2011].
[44] Samwald, M., Jentzsch, A., Bouton, C., et al, Linked open drug data for pharmaceutical research and development. Journal of Cheminformatics 2011; 3:19, doi: 10.1186/1758-2946-3-19.
[45] Pistoia Alliance Sequence Services Project. http://www.pistoiaalliance.org/workinggroups/sequence-services.html. Accessed 14 September 2011.
[46] OpenPhacts Open Pharmacological Space. http://www.openphacts.org/. Accessed 30 August 2011.
[47] Homepage of PostgreSQL. http://www.postgresql.org/. Accessed 30 August 2011.
[48] PostgreSQL featured users. http://www.postgresql.org/about/users. Accessed 30 August 2011.
[49] Homepage of Mysql. http://mysql.com/. Accessed 30 August 2011.
[50] GGA Open-Source Initiative: Products, Ketcher. http://ggasoftware.com/opensource/ketcher. Accessed 30 August 2011.
[51] RDKit: Cheminformatics and Machine Learning Software. http://rdkit.org/. Accessed 30 August 2011.