The economics of free/open source software in industry
Abstract:
Free and open source software has many attractive qualities, perhaps none more so than the price tag. However, does 'free' really mean free? In this chapter, I consider the process of implementing FLOSS systems within an enterprise environment. I highlight the hidden costs of such deployments that must be considered and contrasted with commercial alternatives. I also describe potential business models that would support the adoption of FLOSS within industry by providing support, training and bespoke customisation. Finally, the role of pre-competitive initiatives and their relevance to supporting open source initiatives is presented.
22.1 Introduction
In the last few years use of free/libre open source (FLOSS) software has exploded. Versions of Linux are found in millions of television set-top boxes, Google's Android is arguably the leading smart phone operating system, countless websites are powered by Apache, and millions of Blogs by WordPress. However, for total number of users Mozilla's Firefox browser is hard to beat, with an estimated 270 million active users (as measured by active security pings received by Mozilla [1]), accounting for around 20% of web traffic in general surveys of such things. Firefox is easy to use, highly standards-compliant, secure, well supported, easy to extend with an ecosystem of thousands of plug-ins, and of course it is free to download, use or distribute.
It therefore seemed a perfectly reasonable request when, at a department town-hall meeting in 2009, Jim Finkle asked the US Secretary of State, Hilary Clinton [2] 'Can you please let the staff use an alternative web browser called Firefox?'. The reply, which came from under-secretary Kennedy, that '. . It is an expense question . .' was interrupted from the audience with a shout of 'it's free'. Kennedy continued 'Nothing is free. It's a question of the resources to manage multiple systems.…'. He is, of course, absolutely right, that in a large organisation, be that governmental, academic or corporate there are many hidden costs in rolling out, securing, insuring, maintaining and supporting any software. These costs are incurred, regardless of whether the software is open or closed source, free or expensive.
In the following chapter, I will explore some of these costs in more detail, and analyse a real-world example of how parts of the life sciences industry have come together to try and find new ways they can use open source software in the fight against disease. Before delving into the practical use within industry, I will provide an introduction to the technologies and massive data volumes under consideration by exploring recent advances in human genetics.
22.2 Background
22.2.1 The Human Genome Project
The Human Genome Project is in many ways one of the wonders of the modern world. Thousands of scientists from laboratories around the globe spent a decade and $3 billion to produce what then US President Bill Clinton described as '… without a doubt this is the most important, most wondrous map, ever produced by humankind' [3]. There was great excitement and anticipation that its publication would herald a new era of medical breakthroughs, and it would only be a matter of time until diseases like cancer would only be heard about in history books. I do not intend to go into detail about how the Human Genome Project was delivered, if you are curious its Wikipedia page [4] is an excellent starting point. It is, however, worth mentioning that the human genome is essentially an encyclopaedia written in 46 volumes (known as chromosomes), and written in the language of DNA. The language of DNA is made up of an alphabet of four letters, rather than the 26 we use in English. These DNA letters are called 'base pairs'. The human genome is about 3 billion DNA base pairs in length. This makes the maths of working out the cost of sequencing each base pair a remarkably simple affair of dividing the total $3 billion project cost by 3 billion, giving us a dollar per base pair. Given that each base pair can be coded by 2 bits of information then the whole genome could be stored on about 750 MB of disk space. The cost of storing this much information, even 10 years ago would only be a few dollars, making the data storage costs a microscopic percentage of the overall project costs.
22.2.2 Things get a little more complex
There were huge celebrations in the scientific community when the human genome was published, but this was really only the beginning of the work that needed to be done to get value from the genome. The next step was to read and understand the information in the 46 volumes. Fortunately, just like in an encyclopaedia the genome is broken down into a number of entries or articles. In the world of genetics these entries are called genes. As humans are quite complicated animals, it was expected that we would have a large number of genes, many more than simpler organisms such as worms, flies, plants, etc. All we had to do was work out which gene corresponded to a given human trait and we would have unlocked the secrets to human biology, with hopefully fantastic benefits to healthcare. Unfortunately, there was a problem, it turned out that there were far fewer genes in humans than originally expected. The total number of genes in the human genome was found to be around 23 000. To put this in perspective that is about 9000 genes less than the corn that grows in our fields has, and only just 7000 more than a very simple worm.
As scientists investigated further, new levels of complexity were discovered. If we keep with the analogy of the genome as an encyclopaedia, genes can be thought of as the topics within it. Researchers discovered that paragraphs within each topic could be read or ignored, depending on circumstances, which would change the meaning of the article (this is known as splice variants in genetics). They also found many spelling changes, small additions or omissions from one person's genome to the next. It soon became apparent that these differences were the key to unlocking many more secrets of the human genome. Initially it was only possible to investigate these differences on a very small scale and on very small parts of the genome. However, in the last couple of years huge advances have been made in the science of DNA sequencing; collectively called NGS (next-generation sequencing [5]).
There are several types of NGS technology, and heavy innovation and competition in the area is driving down sequencing costs rapidly. Most NGS technology relies on a process known as 'shotgun' sequencing. In simple terms, the DNA to be sequenced is broken up into millions of small pieces. Through a number of innovative processes, the sequences of these short pieces of DNA are derived. However, these overlapping short 'reads' must be re-assembled into the correct order, to produce a final full-length sequence of DNA. If each of the short sequences is 100 bp long, that would mean an entire human genome could be constructed from 30 million of these. However, to successfully perform the assembly, overlapping sequences are required, allowing gradual extension of the master DNA sequence. Thus, at least one more genomes' worth of short sequences is required, and in practice this is usually somewhere between five and ten genome's worth – known as the coverage level. At ten-fold coverage, there would be a total of 300 000 000 short sequences or 30 000 000 000 base pairs to align. Even that is not the end of the story, each of those 30 billion base pairs has confidence data attached to it; a statistical assessment of whether the sequencing machine was able to identify the base pair correctly. This information is critical in shielding the assembly process from false variations that are due to the sequencing process rather than real human genetic variation. For an informatician, the result of all of this is that each sample run through the NGS procedure generates a vast amount of data (~ 100 GB), ultimately creating experimental data sets in excess of TB of data. Efficient storage and processing of these data is of great importance to academic and industrial researchers alike (see also Chapter 10 by Holdstock and Chapter 11 by Burrell and MacLean for other perspectives).
22.3 Open source innovation
Many of the large projects that are starting to tackle the challenge of unravelling human genetic variation are publicly funded. A great example of such an effort is the '1000 genomes project' [6], which is looking to sequence the genomes of well over a thousand people from many ethnic backgrounds in order to give humanity its best insight yet into the genetic variation of our species. Collecting the samples, preparing them and running them through the NGS platforms, has been a huge logistics and project management operation. However, the part of the project of most relevance here is the informatics task of managing this data avalanche. In particular, analysing and assembling the results into useful information that biologists are able mine and use to generate new insights into human variation. A project of this scale had never been attempted previously in the world of biology, new technology has been developed and iteratively improved by some of the world's most prestigious research institutes. As many of these institutes receive public funding, there has been a long tradition of publishing their software under open source licences. This has many benefits.
![]() By definition open source code has nothing hidden, so its function is completely transparent and easy to examine and analyse. This allows for scientific peer review of a method, easy repetition of experiments and for scientific debate about the best way to achieve a given goal.
By definition open source code has nothing hidden, so its function is completely transparent and easy to examine and analyse. This allows for scientific peer review of a method, easy repetition of experiments and for scientific debate about the best way to achieve a given goal.
![]() This transparency allows other scientific groups not only to replicate experiments, but also to add new features, or indeed suggest code improvements without fear of a team of lawyers knocking on their door.
This transparency allows other scientific groups not only to replicate experiments, but also to add new features, or indeed suggest code improvements without fear of a team of lawyers knocking on their door.
![]() A project like the 1000 genomes is for the benefit of all mankind and as soon as the data are available in a suitable format, they are put up on an FTP site for anyone in the world to download. There are no commercial considerations, no intellectual property to collect licence fees on, no shareholders looking for a return on investment. Everything that is done is available to everyone.
A project like the 1000 genomes is for the benefit of all mankind and as soon as the data are available in a suitable format, they are put up on an FTP site for anyone in the world to download. There are no commercial considerations, no intellectual property to collect licence fees on, no shareholders looking for a return on investment. Everything that is done is available to everyone.
Often open/community approaches can be criticised, rightly or wrongly, for not being as 'good', well supported or feature-rich as their commercial rivals. However, the sheer scale of investment and intellect that is focused on challenges such as the 1000 genomes produces science of the very highest quality. Software that has been built and tested against the biggest and most demanding data sets in history, and consequently really sets a gold standard. The technology developed for projects such as the 1000 genomes is being re-used in many additional projects; from helping sub-Saharan farmers by understanding the relationship of genetics to viability of their cattle, to helping answer questions about the nature of human migration out of Africa some 10 000 years ago. However, by far and away the biggest value of the utilisation of this technology will be in the battle against disease.
22.4 Open source software in the pharmaceutical industry
It takes teams of thousands of scientists, billions of dollars and many years of hard work to produce a new medicine. The risks are huge, and competition between pharmaceutical companies is intense, as there are no prizes for being second to patenting the same molecule. As a consequence, pharmaceutical companies are as security-conscious as those in the defence industry. Security is not the only concern for IT departments in the pharmaceutical industry. As soon as a potential new pharmaceutical product reaches clinical testing, there are a huge set of rules that must be followed to ensure that everything is done reproducibly and correctly. A huge amount of effort is put into validating these systems (see Chapter 21 by Stokes), which would need to be re-done at great expense if things change. This means that there must be an extensive support network behind these systems, and with that appropriate contacts and guarantees.
An IT manager is much more confident of getting support from a large software company, which offers teams of experts and global support for its software for a decade or more, than the hope that someone on a support Wiki might be able to offer a suggestion when something goes wrong. These large software companies have strong patent protection on their software, which enables them to make money from their work. This enables them to grow into global corporations, capable of offering 24/7 support teams of experts in many languages, and legal protection of millions of dollars to their customers if data were to leak or service be interrupted. As such, the pharmaceutical industry has generally been slow to adopt open source software, simply because it does not offer the protections and support guarantees of a commercial counterpart. However, where those protections do exist (for example, from RedHat for its version of Linux), open source software can quickly appear in the data centres and scientific workstations of pharmaceutical companies.
22.5 Open source as a catalyst for pre-competitive collaboration in the pharmaceutical industry
The past few pages have outlined how the pharmaceutical industry has recently found itself pulled in two contradictory directions. On the one hand there is the need for a reliable, supported, secure suite of software tools, and on the other is a wealth of high-quality, powerful open source software written to analyse and manage the torrent of data produced by the latest DNA sequencing technology. Traditionally when faced with this type of challenge, pharmaceutical companies would turn to their vast internal IT departments, who would internally develop the skills needed to manage and support the software. However, this is an expensive and wasteful approach, with each pharmaceutical company having to build an internal platform, integrate it with existing systems and provide the high level of expert support demanded by the scientists in their company. The skills needed to build such a platform are a blend of world-class IT with cutting-edge scientific knowledge, which is rare, valuable and in high demand, which in turn makes it expensive.
However, in many instances, even when pharmaceutical companies do make significant internal investments in building and supporting IT, it is arguable that they provide any competitive advantage over their peers [7]. This is particularly true in the DNA sequence domain, where many of the core algorithms and software are free and open for anyone to use. It has quickly become apparent that the competitive value between companies came from the results generated by these systems and not from building the systems themselves. It is from this type of realisation that organisations such as the Pistoia Alliance [8] were conceived.
The Pistoia Alliance is a non-profit organisation that was set up in recognition of the great deal of important, yet pre-competitive work being done in the life sciences industry. The Pistoia Alliance has members ranging from giant Fortune 500 companies, through to new start-ups, and it spans both life sciences companies, and IT vendors who supply all sorts of the infrastructure used in modern pharmaceutical research. The stated goal of the Pistoia Alliance is to 'lower the barriers to innovation in Life-sciences research'. One excellent way of achieving this is to develop, champion and adopt open standards or technologies that make it easy to transfer data and collaborate with other companies, institutions and researchers. The Pistoia Alliance has been running several projects ranging from open standards for transferring scientific observations from electronic lab notebooks, to developing new open technology to allow better queries across published scientific literature. However, the project that illustrates how industry can work in a more open environment in the area of DNA sequence technology is known as 'Sequence Services'.
22.6 The Pistoia Alliance Sequence Services Project
NGS has produced a boom in commercial opportunities; from companies that build the robots to do the work, and reagents needed to fuel the process, to high-tech sequencing centres with the capacity and expertise to service the sequencing needs of many life science companies. Consequently, a great deal of sequencing is outsourced to these centres. Once the raw sequencing has been completed, the complex process of assembling the DNA jigsaw starts, prior to the next level of analysis where patterns in variation are identified. These are correlated with other observable traits (know as phenotypes to geneticists) such as how well someone in a clinical trial responds to the treatment. Increasingly, it is likely that some or all of this analysis may be done by specialist companies, external experts or via research agreements with universities. All of this work is done under complex legal contracts and conditions, which ensure appropriately high ethical and security standards are always adhered to.
Managing these complex collaborations can produce a huge logistical and security challenge for IT departments. The first of these problems is in shipping the data to where they are needed. As the data sets tend to be very large, sending the data over the internet (usually a process known as Secure File Transfer Protocol, or SFTP) can take a very long time. So long, that it is often far faster to use what is jokingly called the 'sneaker net', which in reality tends to be an overnight courier service transporting encrypted hard drives. This then involves the complexity of getting the decryption keys to the recipients of the hard drives, and therefore the setting up of a key server infrastructure. Once the data have been decrypted, they inevitably get moved onto a set of servers where they can be assembled and analysed. Controlling who has access to the data, and managing copies becomes hugely complex, and expensive to manage.
The past few years has seen a new phrase enter the IT professional's vocabulary, 'cloud computing'. By 2011 almost every major IT company offers its own cloud platform, and hundreds of analysts' reports recommend that corporations leverage the cloud as part of their IT infrastructure strategies. In theory, the cloud was the perfect platform to answer the challenge of NGS data. The cloud is fantastically scalable, so IT managers would not have to estimate data volumes at the beginning of a budget cycle, then have spare capacity for most of the year, only to have to scramble for more at the end of the cycle if use had been underestimated. The cloud also has huge on-demand computational resources, again meaning you only pay for the cycles you use, not for a supercomputer that sits idle for 80% of the year. The cloud also sits outside a corporation's data centres, which is excellent for supporting collaboration with third parties, and prevents corporate fire walls having holes punched through them to allow third-party access. It is also expected that the sheer scale of cloud operations should offer considerable cost-savings over replicating the same functionality in a private data centre. In fact, open source software lends itself very nicely to cloud deployment. First, there are no licencing issues about running the software on hardware that is outside of an organisation, and, second, there are no restrictions about the number of CPUs that can accommodate the workload (other than justifying the actual running costs of course). In contrast, there are often restrictions placed on such things with commercial software depending on individual business models.
There are, however, also risks in using open source software especially as part of a commercial process. For open source software to be a key part of such a process the following questions would need to be addressed.
1. Is the software written in a way that prevents malicious use, in a nutshell is it secure and hack-resistant?
2. How would a scientist get support? As part of a key platform support has to be more than posting a question on a wiki and hoping someone responds in a reasonable time with the right answer. Also in a modern global pharmaceutical company these scientists could be in any time zone.
3. How would users get training? Again this is expected to be more than a FAQ sheet, and depending on the software could range from instructor-led classes, through to interactive online training.
4. How would the system receive updates and upgrades? How quickly would this follow release? How would it be tested? What are the change control mechanisms?
5. Are there any restrictions or incompatibilities with the various open source licences, and the use of other software and services that may restrict how things like extra security (e.g. user authentication) can be wrapped round an open source package?
It was with these questions in mind that the Pistoia Sequence Services Project was launched. The ultimate aim was to host DNA sequence software in the cloud, learning what works and addressing some of the questions above. Indeed, two of the above questions were quickly resolved by the 'vendor supported' model that Pistoia is exploring. First, for question 2, commercial vendors would agree to host open source software and use their current support solutions to address the issues of their industry users. In particular, ensuring that an expert inside their organisations would always be able to respond to support requests. In terms of question 3 concerning training, the Sequence Services Project selected software to host that already had excellent online training modules that had been written by the public institute that wrote the software. Finally, question 4, the challenge of updating the software and its underlying data when a new production version was released, is also not insurmountable. Indeed, a one month lag from public release to hosted service is perfectly acceptable while whatever testing was undertaken by the hosting provider. Thus, the vendor-hosted model of free/open source software addresses some of the key initial challenges to industry adoption. However, the project's aim was to go further and investigate the remaining issues.
Perhaps surprisingly, there can be some challenges to the adoption of open source software in a commercial environment due to the licence terms that are used when releasing software. Take the following statement which has been taken from an open source licence (but anonymised).
You agree that you will deliver, and you will cause all sublicensees to deliver, to XXX copies of all Modifications that are made to the Software, on or before the date on which the Modifications are distributed to any third party, and you hereby grant to XXX, and will cause all of your sublicensees to grant to XXX, a non-exclusive, royalty-free, irrevocable, non-terminable license to use, reproduce, make derivative works of, display, perform, and distribute any and all Modifications in source code and binary code form for any purpose and to sublicense such rights to others.
From the point of view of the author of the software, it is not unreasonable to expect people who are using and benefiting from your hard work to contribute back any improvements they add. However, this may mean that the software could not be easily used with any commercial tools. It may be completely reasonable for a company to want to put some user authentication process into the software, or perhaps a plug-in that allows the use of commercial visualisation software. At the very minimum, lawyers now need to be involved (probably on both sides) to agree what constitutes a 'modification', and even if a third-party developer could be commissioned by a company to do any work on the code without forcing the original author into the development process (and into a confidentiality agreement).
There are a great number of open source licences available, but they are certainly not all the same, and indeed not always compatible with each other. For example, when the open source 'SpamAssassin' software was moved into Apache, project organisers spent months getting permission for the move from all the licence holders (around 100 in total) [9]. Not all contributors could be tracked down, and some software had to be rewritten to allow it all to move to the Apache licence. This demonstrates how important it is to understand any reach through claims and restrictions that are associated with a given open source licence. In a large company this would almost always involve taking legal advice, and producing rules on how to use the software safely within the licence; another huge potential hidden cost of using open source software. For anyone who wants to know more about the many and various open source licences an excellent resource from the 'Open Source Software Advisory Service' is available [10] and the subject was covered in a recent book chapter [11].
One of the biggest challenges in deploying open source software in a commercial environment is ensuring that it is secure. Each year there are a huge number of malicious hackings of corporations, with many well-known companies suffering significant damage to their businesses and reputations. The IT vendors who joined the sequence services' team all knew how to build secure platforms. Many work closely with customers in the financial and defence industries, and are consequently experts in building secure IT platforms. When building these systems, security is thought about and tested at every step. As open source software is often not built with commercial considerations, the developers quite rightly primarily focus on the functional aspects of the software. Although there is some highly secure open source software (such as EnGarde Linux), often the developer's focus is not necessarily securing the software from skilled 'blackhat' hackers and crackers intent on breaking in. Traditionally this has not been a huge problem. Where the software is hosted and available on the internet, it either contains no data or only already publicly available data. Given that the code is already open source, there is little incentive for anyone to hack into the system other than pure vandalism, and this means most sites come under nothing more than basic 'script kiddie' attack. Where a large commercial company uses the software it would be normal for it to be hosted inside their own data centres on the internal intranet, which is safely protected from the external world by large, sophisticated corporate firewalls.
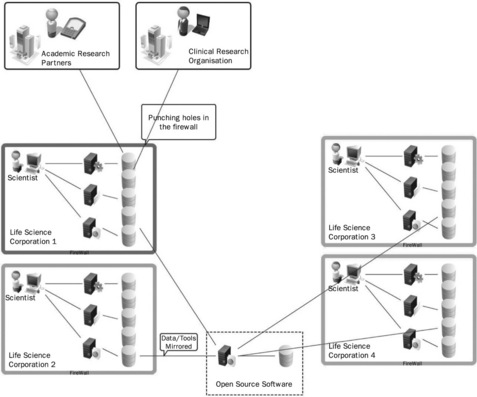
This intranet solution (shown in Figure 22.1) does, of course, have a number of drawbacks. First, there is the cost involved of hosting the software, testing the software and managing updates, as well as providing training to the IT helpdesk, and the users. These costs are not insignificant, and in many cases make open source software as expensive to run and support as commercial alternatives, especially when training material of a high standard is not already available. This cost is then duplicated many times at other corporations that are doing similar research. On top of that, as many companies begin to work in more collaborative ways, they open B2B (business to business) connections through their firewalls. This has led to more concern about the security vulnerabilities of the software hosted internally.
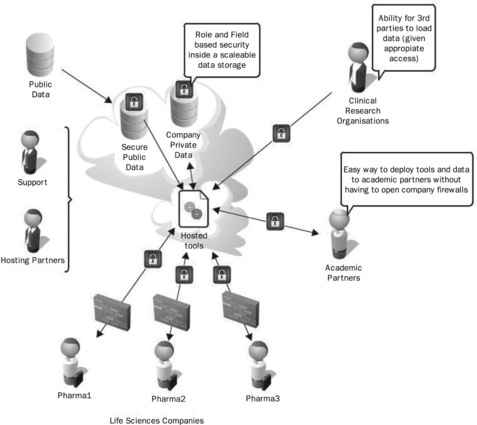
The vision of the Sequence Services Project is to try to reduce the total costs of running and maintaining such a system, while ensuring that functionality, performance and, above all, security are not compromised. This new vision is shown in Figure 22.2, where the services are hosted and maintained by a third-party vendor.
Pistoia chose an open source suite of software that is well known in the bioinformatics world as its test software for the proof of concept work. Before starting there were discussions with the institute that wrote the software, building an agreement to communicate the results of the project and, in particular, share any recommendations or security vulnerabilities that were discovered.
In order to test and maintain security a number of approaches were taken. The first was to run an extensive ethical hack of the chosen open source software. Pistoia did this by employing the professional services of a global IT company with a specialist security division, producing an extensive report containing a breakdown of vulnerabilities. Although I cannot list specific security issues found in the Pistoia project, I would like to highlight the most common vulnerabilities found in such software. For those that are particularly interested in this topic, a valuable resource in this area plus a list of the current top 10 vulnerabilities can be found at the Open Web Application Security Project (OWASP) website [12].
![]() Application allows uploading of malware
Application allows uploading of malware
Whenever a file is being uploaded, at minimum the software should check if this is the type of file it is expecting, and stop all others. A further step could be taken by running a virus check on the file.
When a user has authenticated and gained access to the system there should be a method to log out if the user has been inactive for a set period of time.
A common example of this is the ability once you have been given access to a system to be able to find access areas that you shouldn't by manually entering a URL.
This is when an attacker can use a bug to re-direct a user to a website that the attacker controls. This will usually be made to look like the original site, and can be used to harvest information like login IDs and passwords.
The system should not offer to remember passwords or similar security tokens.
![]() Web server directory indexing enabled
Web server directory indexing enabled
This is often left on by default, and can give an attacker some indication of where weaknesses exist or where sensitive data may reside.
![]() Sensitive information disclosed in URL
Sensitive information disclosed in URL
Some web-hosted software will place certain pieces of information, such as search terms, in a URL (. . . . search = web + site + security + . . .). This could be viewed by an attacker if the connection is not secured.
An error message should ideally be written so it is clear to the user an error has occurred; however, it should not contain any information that an attacker may find useful (such as the user's login ID, software versions, etc.) This can all be put in an error log and viewed securely by a support team.
This is a technique whereby an attacker can run a database search that the software would not normally allow, by manipulating the input to the application. This is not difficult to prevent if developers are thinking about security, so that the system only ever runs validated input that it is expecting.
Users should not be able to log into the system more than once at any given time.
![]() Web server advertises version information in headers
Web server advertises version information in headers
Revealing the version of the server software currently running makes it easier for hackers to search for vulnerabilities for that version.
Even if a system has been found to have no known vulnerabilities during testing, there are further steps that can to be taken to minimise any risks.
![]() Regular security re-testing, at least at each version release – this, of course, costs money to maintain.
Regular security re-testing, at least at each version release – this, of course, costs money to maintain.
![]() Use of virtual private clouds, where private data are clearly segregated.
Use of virtual private clouds, where private data are clearly segregated.
![]() Use of IP filtering at the firewall to reject any IP address other than the approved ones. Although IP addresses can be spoofed, a hacker would need to know the correct ones to spoof.
Use of IP filtering at the firewall to reject any IP address other than the approved ones. Although IP addresses can be spoofed, a hacker would need to know the correct ones to spoof.
![]() Use of appropriate validated authentication standards, obviating the need for services to provide their own ad hoc systems.
Use of appropriate validated authentication standards, obviating the need for services to provide their own ad hoc systems.
![]() Use of remote encryption key servers, which would prevent even the vendor hosting the software from being able to view the private data they are hosting.
Use of remote encryption key servers, which would prevent even the vendor hosting the software from being able to view the private data they are hosting.
![]() Use of two factor authentication. Many users will use the same short password for every service, meaning that once an attacker has found a user name and password at one site, they can quickly try it at many others. Two factor authentication usually takes the form of a device that can give a seemingly random number that changes on a regular basis which would need to be entered along with the traditional user name and password.
Use of two factor authentication. Many users will use the same short password for every service, meaning that once an attacker has found a user name and password at one site, they can quickly try it at many others. Two factor authentication usually takes the form of a device that can give a seemingly random number that changes on a regular basis which would need to be entered along with the traditional user name and password.
![]() The use of appropriate insurance to provide some financial compensation if a service is down or breached.
The use of appropriate insurance to provide some financial compensation if a service is down or breached.
22.7 Conclusion
As I hope I have highlighted, just as under-secretary Kennedy explained in his answer at the beginning of this chapter, there are many costs associated with the use of open source software in government departments or in industry. As soon as there is a need to store or process sensitive data such as medical records, then the costs and complexity can increase considerably. The cost-benefits of open source software being free to download and use can quickly be lost once training, support, security, legal reviews, update management, insurance and so on are taken into account.
However, in many areas of scientific research, open source software projects are truly world leading. For industry to get maximum value from open source software it needs to actively participate in the development process. Organisations like Pistoia show one way that this participation can be done, where many companies come together in areas that are important (yet pre-competitive). This approach has many benefits.
![]() Shared requirements – allowing industry to speak with one voice, rather than many different ones. This can obviously help with understanding how software might be used, in prioritising future work and in improving and adopting open standards.
Shared requirements – allowing industry to speak with one voice, rather than many different ones. This can obviously help with understanding how software might be used, in prioritising future work and in improving and adopting open standards.
![]() Shared costs and risks – with many companies all sharing the cost of things like expert security reviews of software, the share of the cost to each company can quickly become very reasonable.
Shared costs and risks – with many companies all sharing the cost of things like expert security reviews of software, the share of the cost to each company can quickly become very reasonable.
![]() Feedback and collaboration – even if industry is not participating directly with the writing of a given open source software package, it can help to build relationships with the authors. This way the authors can better understand industry requirements. Importantly, authors will not see a list of security vulnerabilities merely as a criticism, but rather as a contribution to making the software better for everyone.
Feedback and collaboration – even if industry is not participating directly with the writing of a given open source software package, it can help to build relationships with the authors. This way the authors can better understand industry requirements. Importantly, authors will not see a list of security vulnerabilities merely as a criticism, but rather as a contribution to making the software better for everyone.
Open source software is very likely to play an important role in the life sciences industry in the future, with companies not only using open source software, but also actively contributing back to the community. Indeed, earlier in this book (Chapter 1) we saw a great example of exactly this from Claus Stie Kallesøe's description of the LSP4All software. Another exciting example of industry using open source software to help drive innovation is demonstrated by the Pistoia Alliance's recent 'Sequence Squeeze' competition. The Pistoia Alliance advertised a challenge to identify improved algorithms to compress the huge volumes of data produced by NGS. In order to find an answer they reached out to the world by offering a $15 000 prize to the best new algorithm. All algorithms had to be submitted via the sourceforge website, and under the BSD2 licence. The BSD2 licence was deliberately chosen by Pistoia as it has minimal requirements about how the software can be used and re-distributed – there are no requirements on users to share back any modifications (although, obviously, it would be nice if they did). This should lower the legal barriers within companies to adopting the software, and is a great example of industry embracing open source standards to help drive innovation. The challenge was open to anyone and entries were ranked on various statistics concerning data compression and performance. A detailed breakdown of the results of the challenge is available from [13].
In conclusion, it is clear that free and open source software is not really 'cost-free' in an industrial setting. It is also true that there can be issues that make deployment difficult, particularly where sensitive data are concerned. However, by developing new models for hosting and supporting such software we could be witnessing a new direction for the use of free and open source software in commercial environments.
22.8 References
[1] http://weblogs.mozillazine.orglasalarchivesl2009I0SIfirefox_at_270.html
[2] http://www.state.govlsecretarylrml2009aljulyll2S949.htm.
[3] http://www.dnalc.orglviewl1S073-Completion-of-a-draft-of-the-human-genome-Bill-Clinton.html.
[4] http://en.wikipedia.orglwikilHuman_Genome_Project.
[5] http://en.wikipedia.orglwikilDNA_sequencing.
[6] The 1000 Genomes Project Consortium, A map of human genome variation from population-scale sequencing. Nature. 2010; 467:1061–1073.
[7] Barnes, M.R., et al. Lowering industry firewalls: pre-competitive informatics initiatives in drug discovery. Nature Reviews Drug Discovery. 2009; 8:701–708.
[8] http://www.pistoiaalliance.org.
[9] http://en.wikipedia.orglwikilSpamAssassin.
[10] http://www.OSS-watch.ac.uk.
[11] Wilbanks, J. Intellectual Property Aspects Of Collaboration in Collaborative Computational Technologies for Biomedical Research (Ekins S, Hupcey MAZ, Williams AJ eds.) Wiley (London) ISBN0470638036.