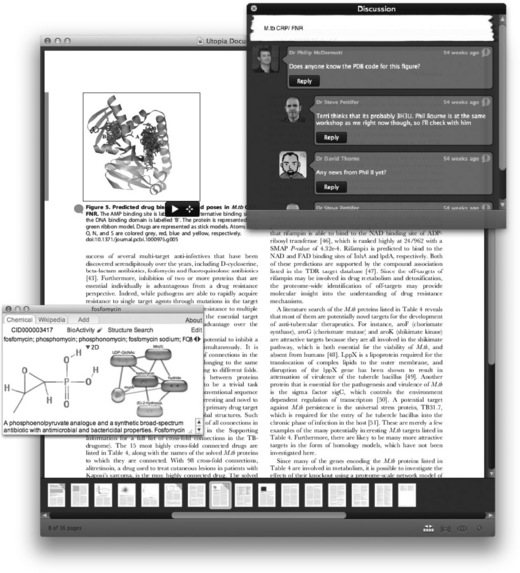
Utopia Documents: transforming how industrial scientists interact with the scientific literature
Abstract:
The number of articles published in the scientific literature continues to grow at an alarming rate, making it increasingly difficult for researchers to find key facts. Although electronic publications and libraries make it relatively easy to access articles, working out which ones are important, how their content relates to the growing number of online databases and resources, and making this information easily available to colleagues remains challenging. We present Utopia Documents, a visualization tool that provides a fresh perspective on the scientific literature.
It is a curious fact that, throughout history, man’s capacity to create knowledge has far outstripped his ability to disseminate, assimilate, and exploit that knowledge effectively. Until recently, the bottleneck was very much a physical one, with hand-written scrolls, and then printed manuscripts and books, being deposited in libraries to which access was limited, either by politics, social factors, or simply the need to physically travel to a particular location in order to access the content. Digital technologies have not solved the problem – and, it could be argued, have in some ways made matters worse.
Although it is no longer necessary to visit a library in person to read a scholarly work, so much material now exists that making sense of what is relevant is becoming increasingly difficult. In the life sciences alone, a new article is published every two minutes [1], and the number of articles has been growing exponentially at around 4% per year for the past 20 years [2]. In 2011, for example, over 14 000 papers were published relating in some way to Human Immunodeficiency Virus (admittedly, this is a much broader topic than any one researcher would care to explore in detail; nevertheless, it gives some indication of the scale of the problem) [3]. If we add to the peer-reviewed literature all the information that is deposited in databases, or written informally on personal blogs, matters get more complex still. The problem has been characterized, at a broad level, as being one of ‘filter failure’ [4], that is it’s not that there is too much information per se – as, surely, lots of information has to be a good thing, and there has always been more available than we are able to consume – but, rather, that we are lacking mechanisms to convert information into meaningful knowledge, to find what is relevant to our particular needs, and to make connections between the disjoint data.
The problem is particularly acute in the life sciences. As pharmaceutical companies race to find new drugs, it has become clear that mastery of the literature is at least as important as mastery of the test tube or the sequencing machine. It has been documented that some failures of pharmaceutical company projects – which are costly on a mind-boggling scale, and typically result in failure – could have been avoided if scientists within the company had simply been able to find the relevant scientific article beforehand [5]. This inability to find the ‘right article’ is not for the want of trying: pharmaceutical companies employ large teams of scientists to scour the literature for just such information; it is simply that, even with the best tools and the most proficient scientists, there is more information ‘out there’ than can currently be processed in a timely way.
The danger of characterizing this as simple ‘filter failure’ is that it makes the solution sound trivial: get a better ‘filter’, and the problem goes away. The difficulty, of course, is that no filter yet exists that is sophisticated enough to cope with the complexities of life science data and the way in which it is described in ‘human-readable’ form. The simple key word searches or faceted browsing techniques that work so well for buying consumer goods or for booking a holiday online, break down quickly when faced with complex, context-sensitive, inter-related, incomplete and often ambiguous scientific data and articles. In many cases information gleaned from articles is already known in some part of an organization, but its relevance to some other new project is not discovered until after the fact. For organizations such as pharmaceutical companies, as the volume of information grows, the problem of ‘knowing what we already know’ is set to become worse [6].
At the heart of this problem lies a simple truth. We are now completely reliant on machines to help process the knowledge we generate; but humans and machines speak radically different languages. To perform efficiently, machines require formal, unambiguous and rigorous descriptions of concepts, captured in machine-readable form. Humans, on the other hand, need the freedom to write ‘natural language’, with all its associated ambiguities, nuances and subtleties, in order to set down and disseminate ideas. The trouble is, other than trained ‘ontologists’, mathematicians, and logicians, very few scientists can turn complex thoughts into the kind of formal, mathematical representations that can be manipulated computationally; and very few ontologists, mathematicians, and logicians understand biology and chemistry in enough detail to capture leading-edge thinking in the pharmaceutical or life sciences. For the time being, at least, we are faced with the challenge of bridging this divide.
Text- and data-mining techniques have made some progress here, attempting to extract meaningful assertions from written prose, and to capture these in machine-readable form (e.g. [7]). But this is far from being a solved problem – the vagaries of natural-language processing, coupled with the complex relationships between the terminology used in the life sciences and records in biomedical databases make this a non-trivial task.
In recent years, in what we consider to be a frustrated and frustrating distraction from the real issues, many have pointed the finger of blame at the file format used by publishers to distribute scientific articles: Adobe’s PDF. It has variously been described as ‘an insult to science’, ‘antithetical to the spirit of the web’, and like ‘building a telephone and then using it to transmit Morse Code’, as though somehow the format itself is responsible for preventing machines from accessing its content [8]. Although it is true that extracting text from PDFs is a slightly more unwieldy process than it is from some other formats, such as XHTML, to accuse the PDF as the culprit is to entirely miss the point: the real problem arises from the gulf between human- and machine-readable language, not merely from the file-format in which this natural language is being stored. It is worth setting the record straight on two PDF-related facts.
1. Although originating in a commercial environment, PDF is now – and has been for some considerable time – an open format. The specifications of the PDF have been publically available for many years, and have been formally ‘public domain’ since 2008, when Adobe released the format as ISO 32000–1 and officially waived any patents relating to creating or consuming PDF content [9]. It is, however, undoubtedly a more verbose, and arguably less-elegant format than more recent offerings.
2. The PDF has the potential to be as ‘semantically rich’ as any of the other alternatives (e.g. eBook, XHTML), and, since version 1.4 in 2001, has had the facility to include arbitrarily sophisticated meta-data. The fact that virtually no publishers use this feature is not a fault of the PDF, and such semantic richness is just as conspicuously absent from the other formats used to publish scientific material. Speculation as to which link in the chain between author, publisher, and consumer causes this omission, we leave entirely to the reader.
Regardless of whether the PDF is a suitable vehicle for disseminating scientific knowledge, there are two irrefutable facts: first, it is by far the most popular medium by which scientific articles are consumed (accounting for over 80% of downloaded articles); and second, even if publishers move to a more ‘web friendly’ format at some point in the future, vast numbers of legacy articles will remain only as PDFs (many of which are digital scans of back-catalogues that pre-date even this ancient format, some of which have had text extracted via optical character recognition, while many exist only as images) – at the very least, these would require converting into something more ‘semantic’. In spite of significant effort by publishers to enhance their online/HTML content with additional links, meta-data, data and multimedia extensions, the PDF stubbornly remains the format of choice for most readers. The recent growth in modern tools for managing collections of PDFs (e.g. Mendeley [10] and Papers [11]) gives additional backing to the view that the PDF is not in any danger of disappearing yet.
In an attempt to bridge the divide between what scientists actually read, and what computers need to support the process of taming the scientific literature, we embarked on a project to build a new tool for linking the content of articles with research data. The resulting software – Utopia Documents [12, 13] – combines all the advantages of the PDF with the interactivity and interconnectedness of a blog or web page. Acting as an alternative desktop application to Adobe Acrobat and other PDF viewers, Utopia Documents forms two-way links between the traditional human-readable PDF and any available semantic models produced by publishers or the community, and public or in-house data sources. The act of reading an article triggers heuristics that attempt to automatically identify features within the PDF, which can then be semantically annotated, and these annotations published to augment the original article.
The software ‘reads’ a PDF much like a human does, recognizing document content and features (title, authors, key words, sections, figures, etc.) by their typographical layout, and ignoring less-important artifacts and non-document content (such as watermarks and additional information in the header or margins). Having gleaned this information from the PDF, Utopia Documents is then able to automatically link citations and references to online repositories.
As well as creating inbound and outbound links, Utopia Documents incorporates a range of interactive features and visualizations that effectively bring static PDF files to life. For example, the software can turn tables of data into live spreadsheets (Figure 15.1); it can generate charts – already linked to the source data – on the fly; it can turn objects and images (e.g. protein sequences and 3D structures) into live, interactive views; and it can dynamically include data- and text-mining results through customization with appropriate ‘plug-ins’. It also allows readers to add comments to articles, which can then be shared (and commented on) with other users instantaneously, without the need to redistribute the PDF file itself. The system currently links to several major life science and bioinformatics databases, as well as to more general online resources, such as Wikipedia. By means of its plug-in architecture, Utopia Documents can be integrated with any web service-accessible resource, making it easily customizable and extensible. Although it is possible for journal publishers to enhance PDFs with content that Utopia can recognize (noted below), it is important to state that the majority of Utopia’s features work on all modern scientific PDFs, and are not limited to only a few specialized documents.

Figure 15.1 A static PDF table (a) is converted first into an editable ‘spreadsheet’ (b), and then into a dynamic scatterplot (c)
15.1 Utopia Documents in industry
In a commercial context, Utopia Documents aims to provide a new way to enable ‘joined-up thinking’ within institutions, reducing the cost of relating knowledge from within scientific articles to existing in-house and public data, and at the same time supporting auditable interaction with the literature. This results in simplifying the ‘due diligence’ processes essential in modern drug discovery. Other ‘knowledge management’ tools typically provide web-based interfaces that allow scientists to explore their content, but treat the scientific article itself as a distinct and separate entity, somewhat divorced from the discovery process. The consequence of this approach is that scientists are required to repeatedly switch from one application to another while pursuing a line of thinking that requires access both to the literature and to data. Often the situation is worse, and they are forced to print documents out, to scribble notes in the margin and thereby remove the potential for interconnected data completely. Not only does the act of consciously switching require a certain amount of mental inertia (leading inevitably to, ‘Oh, I’ll look that up next time when I’m using Application X’), it interrupts the workflow and thought processes associated with reading the literature. In turn, this makes documenting associations between concepts found in articles and data recorded in other systems difficult and fragmented.
Consider a typical industry use-case: a scientist is reading a paper describing a number of proteins and/or small molecules relevant to a disease or phenotype. Some of these might be new to the reader, who may wish to know a little more about them in order to better understand the findings in the paper. Traditionally, manual copy/pasting of protein and drug names into numerous search engines is the only solution. For compounds, this can also require tedious manual sketching of the molecule into a structure editor. The difficulty in performing what should be very simple tasks often results in either lost time or simply abandoning the enquiry.
Now, consider these same scenarios using Utopia Documents. Simply selecting the protein name using the cursor brings up a rich information panel about that entity (Figure 15.2). Alternatively, text-mining services can be invoked directly from within the Utopia environment to automatically identify interesting entities and relationships, and to link these to in-house data. The result is a highly visual and organized index of the genes, compounds, and diseases mentioned in the text. For compounds, Utopia goes even further, as Figure 15.3 shows. Here, the software has identified a number of small molecules mentioned only by name in the text of the paper. Using (in this example) the ChemSpider [14] API, the actual structures have been retrieved, along with other information, providing a completely new chemical index browser to the paper. Functionality developed as part of the EU Open PHACTS project [15] also allows the retrieval of activity data from public and corporate structure-activity relationship databases.

Figure 15.2 In (a), Utopia Documents shows meta-data relating to the article currently being read; in (b), details of a specific term are displayed, harvested in real time from online data sources
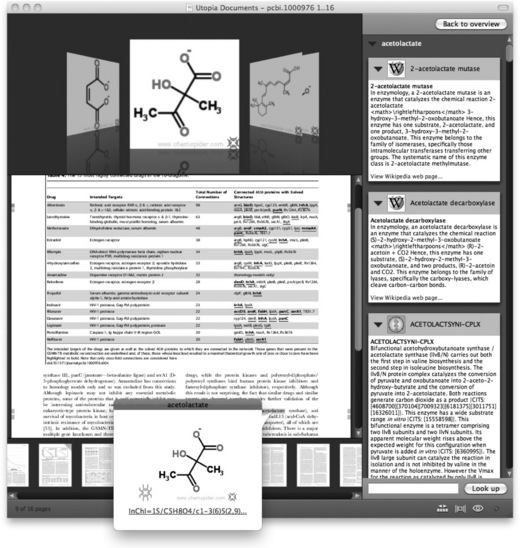
Figure 15.3 A text-mining algorithm has identified chemical entities in the article being read, details of which are displayed in the sidebar and top ‘flow browser’
Finally, because of Utopia’s unique software architecture, many other types of plug-in can be developed, including the ability to interact with tables and to embed applications, such as 3D protein structure viewers, directly in the document. Utopia provides a two-way API, allowing web services to retrieve structured content from the PDF (‘give me all tables from the document’) and inject content into the PDF (‘show these data when the user highlights a compound’). This provides many opportunities to interconnect with internal systems within a company. For instance, highlighting a protein might call for a look-up of the internal project-management system to list which groups are actively researching this target for relevant indications. Similarly, highlighting a compound might invoke a structure-search of the corporate small-molecule database, identifying structurally similar compounds in the internal file. By these means, Utopia connects the content of the PDF into a company’s systems, finally joining one of the most important pieces of the scientific process to the existing information infrastructure.
15.2 Enabling collaboration
Utopia stands apart from other PDF-management tools in the way in which it promotes collaboration and sharing of knowledge within an institution. Most life science industry projects are run by dynamic, ever-changing teams of scientists, with new members being introduced as specialist expertise is required, and scientists typically contributing to many projects at any given time. A researcher faces a constant battle to stay on top of the literature in each project, determining which papers should be read and in what order, and exchanging thoughts with colleagues on the good and bad of what they have digested. Often the solution to both these needs is ad hoc email, reading papers suggested by colleagues and replying with relevant critique. Of course, this relies on the researcher being copied into the email thread and, assuming that this has happened, on finding the email in an ever-growing inbox (in all probability, emailing a published article to a colleague, even within the same organization, is likely to be a technical violation of the license under which the original reader obtained the article).
Utopia changes this by providing a team-based approach to literature management. Unlike other document-management tools, this enables collaboration not just at the level of a paper, but rather, at the level of a paper’s content. Individuals can manage their own paper collections in a familiar and intuitive interface; however, by simply creating team folders and sharing these with colleagues, interesting papers and their related data and annotations can be circulated within relevant groups, without the need to remember to send that email and get everyone’s name right. Simple icons inform team members of new papers in the collection (Figures 15.4 and 15.5). As well as comments, annotations on articles can include data-files (e.g. where the group has re-interpreted a large data set), or links to data held in knowledge-management systems. Thus, anyone reading the paper will see that there is additional information from inside the company that might make a real difference to their interpretation.
15.3 Sharing, while playing by the rules
Unlike other tools, Utopia does not rely on users exchanging specific, annotated copies of PDF articles, or on the storage of PDFs in bespoke institutional document repositories (all of which raise legal questions around the storage and distribution of copyrighted PDFs and the breach of publishers’ licenses). When a user opens a PDF file in Utopia, a small semantic fingerprint is created that uniquely identifies that paper to the system. Any annotations or attachments are associated with this fingerprint and not with the PDF file itself. This has significant consequences for sharing and copyright within corporate environments.
![]() If a scientist on the other side of the world goes to the journal web site and downloads a completely new PDF of a specific article, it has the same fingerprint as any other copy of that article (even if the PDF is watermarked by the digital delivery system to include provenance information). When opened in Utopia, this fingerprint is recognized, and all enhancements and company annotations are retrieved and overlaid on the article in real time. This means that colleagues who may not even know that this paper was of interest to someone in the company are able to exploit the tacit knowledge of the organization that would otherwise be lost.
If a scientist on the other side of the world goes to the journal web site and downloads a completely new PDF of a specific article, it has the same fingerprint as any other copy of that article (even if the PDF is watermarked by the digital delivery system to include provenance information). When opened in Utopia, this fingerprint is recognized, and all enhancements and company annotations are retrieved and overlaid on the article in real time. This means that colleagues who may not even know that this paper was of interest to someone in the company are able to exploit the tacit knowledge of the organization that would otherwise be lost.
![]() A corporate PDF repository is not required. When user A posts a paper to the shared folder, user B sees the title, citation and so on, but does not get a physical copy of the paper until they decide to read that article. Then, a new copy of the PDF is automatically downloaded via the company’s digital delivery service, allowing copyright to be respected and usage/access metrics to be accurately maintained.
A corporate PDF repository is not required. When user A posts a paper to the shared folder, user B sees the title, citation and so on, but does not get a physical copy of the paper until they decide to read that article. Then, a new copy of the PDF is automatically downloaded via the company’s digital delivery service, allowing copyright to be respected and usage/access metrics to be accurately maintained.
15.4 History and future of Utopia Documents
Utopia Documents was originally developed in collaboration with Portland Press Ltd, who have been using the software since December 2009 to enhance the content of their flagship publication, the Biochemical Journal [16]. A publishers’ version of the software allows the journal’s editorial team to ‘endorse’ annotations, confirming the validity of specific associations between terms in articles and particular database entries. This has allowed thorough but rapid markup (typically, 10–15 minutes per article, depending on its suitability for annotation) as part of the normal publishing process. To date, around 1000 articles have been annotated in this way as part of the Semantic Biochemical Journal (http://www.biochemj.org). More recently, the publications of the Royal Society of Chemistry [17] have been augmented by automatically generated annotations on the chemical compounds mentioned in articles, and a plug-in to highlight these in Utopia Documents is included in the latest releases (covering around 50 000 articles, at the time of writing). Other extensions include linking articles automatically to their Altmetric [18] statistics (giving an indication of the popularity of an article based on its citation in blogs, tweets, and other social media), and to online data repositories such as Dryad [19].
Recently, we have engaged in projects with pharmaceutical companies to install the system within an enterprise environment. Utopia consists of two main components, the PDF viewer installed on any user’s PC, and a server, which holds the annotations, fingerprints, and other essential meta-data. Installation of this server behind the corporate firewall provides the security and privacy often required by commercial organizations. In addition, the system will then have access to secure in-house text-mining and data services. Ultimately, this allows commercial entities to use the software without fear of their activities being tracked by competitors, or their sensitive in-house knowledge ‘leaking’ into the public domain. We have explored two major approaches to this configuration: the traditional single installation on a company server, and a cloud-based approach. For the latter, a service provider hosts a secure, tailored instance of the server to which only users from a specific company have access. All of the advantages of an internal installation are preserved, but with much less overhead in maintaining the system. Our experience of the use of Utopia in industry has suggested that the team-based sharing is of greatest initial interest. Industry scientists urgently need tools to help them manage information more effectively, and to manage it as a team.
Deciding on an appropriate license for Utopia Documents has been an interesting and complex endeavor. With the vast majority of its funding so far having come from commercial bodies (primarily publishers and the pharmaceutical industry), rather than from research grants or the public purse, we do not feel the same ethical imperative to make the software open source as we might do had the development been funded through grants derived from public funds. Yet, as academics, we nevertheless find openness extremely attractive. The difficulty arises when the ideals of openness meet the reality of finding sustainable income streams that will allow us to continue the research and development necessary to maintain Utopia Documents as a useful and powerful tool for scientists, whether in commercial or academic environments. What has become clear is that very few of our end-users care about the detailed licensing of the software, as long as the tool itself is robust, reliable, freely available, and has some plausible longevity.
Although it is notionally possible to sustain software development and research on the back of ‘consultancy’ associated with open-source software, there appear to be relatively few concrete examples of where this has worked as a long-term solution for keeping a research team together. Our current model, therefore, is one in which the core software is – and will remain – free to use without restriction (in both academic and commercial settings), while we retain the rights to customize the tool for commercial environments, or to provide specific services to publishers. As academics, finding a model that both sustains our research and provides a freely available resource for the community and ‘the good of science’ has been challenging on many levels; we are nevertheless optimistic that our current approach provides a reasonable balance between long-term sustainability and free availability, and are excited about the potential of Utopia Documents as a means of making more of the scientific literature.
15.5 References
[1] Attwood, T.K., Kell, D.B., Mcdermott, P., Marsh, J., Pettifer, S.R., Thorne, D. Calling international rescue: knowledge lost in literature and data landslide. Biochemical Journal. Dec 2009.
[2] Lu, Z. PubMed and beyond: a survey of web tools for searching bio-medical literature. Database. 2011. [baq036.].
[3] Statistics generated Jan 2012 by searching http://www.ncbi.nlm.nih.gov/pubmed/ for articles mentioning HIV, published in 2011.
[4] http://blip.tv/web2expo/web-2–0-expo-ny-clay-shirky-shirky-com-it-s-not-information-overload-it-s-filter-failure-1283699.
[5] Korstanje, C. Integrated assessment of preclinical data: shifting high attrition rates to earlier phase drug development. Current Opinion in Investigational Drugs. 2003; 4(5):519–521.
[6] Superti-Furga, G., Wieland, F., Cesareni, G. Finally: The digital, democratic age of scientific abstracts. FEBS Letters. 2008; 582(8):1169.
[7] Ananiadou S., McNaught J., eds. Text Mining for Biology and Biomedicine, 2006. [Artech House Books. ISBN 978-1-58053-984-5,].
[8] Pettifer, S., McDermott, P., Marsh, J., Thorne, D., Villeger, A., Attwood, T.K. Ceci n’est pas un hamburger: modelling and representing the scholarly article. Learned Publishing. Jul 2011.
[9] International Organization for Standardization Document management — Portable document format — Part 1: PDF 1.7, IS03200-1:2008.
[10] www.mendeley.com.
[11] http://www.mekentosj.com/papers.
[12] www.utopiadocs.com.
[13] Attwood, T.K., Kell, D.B., Mcdermott, P., Marsh, J., Pettifer, S.R., Thorne, D. Utopia Documents: linking scholarly literature with research data. Bioinformatics. 2010; 26:i540–6.
[14] www.chemspider.com.
[15] www.openphacts.org.
[16] www.biochemj.org.
[17] www.rsc.org.
[18] www.altmetric.com.
[19] www.datadryad.org.