TripleMap: a web-based semantic knowledge discovery and collaboration application for biomedical research
Ola Bildtsen, Mike Hugo, Frans Lawaetz, Erik Bakke, James Hardwick, Nguyen Nguyen, Ted Naleid and Christopher Bouton
Abstract:
TripleMap [1] is a web-based semantic search and knowledge collaboration application for the biomedical research community. Users can register for free and login from anywhere in the world using a standard web browser. TripleMap allows users to search through and analyze the connections in a massive interconnected network of publicly available data being continuously compiled from Linking Open Drug Data [2] sources, the entire corpus of PubMed abstracts and full text articles, more than 10 000 biomedical research relevant RSS feeds, and continuously updating patent literature. TripleMap represents everything in its data network as 'master entities' that integrate all information for any given entity. By searching for, and saving sets of entities, users build, share, and analyze 'dynamic knowledge maps' of entities and their associations.
19.1 The challenge of Big Data
In recent decades computationally based technologies including the internet, mobile devices, inexpensive high-performance workstations, and digital imaging have catalyzed entirely new ways for humans to generate, access, share, and visualize information. As a result, humanity now produces quantities of digital data on an annual basis that far exceed the amount of non-digital data previously generated over the entire span of human history. For example, it is estimated that in 2011 the amount of digital information generated will be 1 800 000 petabytes (PB) or 1.8 zettabytes (ZB) [3]. In comparison:
![]() 5 MB would contain the complete works of Shakespeare [4];
5 MB would contain the complete works of Shakespeare [4];
![]() 10 TB represents the entir printed collection of the US Library of Congress [4];
10 TB represents the entir printed collection of the US Library of Congress [4];
![]() 200 PB contains all printed material ever generated by humanity [4].
200 PB contains all printed material ever generated by humanity [4].
In other words, in 2011 alone, humanity will generate 9000 times more digital information than all of the information ever printed in physical books, magazines, and newspapers. Furthermore, humanity's ability to generate and handle digital data through advances in the means of its generation, transmission, and storage is increasing at an exponential rate [5].
This mind-boggling scale of data is commonly referred to as 'Big Data'. The phrase 'Big Data' is not meant to denote a specific data scale such as 'petabytes' or 'exabytes' (1015 and 1018 bytes, respectively), but instead to be indicative of the ever-increasing sea of data available to humans and the challenges associated with attempting to make sense of it all [6]. The Big Data explosion in the biomedical research sector has been further catalyzed by the completion of the human genome sequencing project [7], the sequencing of genomes from a number of species, and the emergence of a range of correlated, omics and next-generation technologies (e.g. microarrays, next-generation sequencing approaches) as discussed in a number of other chapters in this book.
The availability of Big Data presents humans with tremendous capabilities to 'see the big picture' and gain a 'bird's eye' view of patterns, trends, and interrelationships under consideration and it stands to reason that individuals, teams, and organizations that are able to harness Big Data for searching, analyzing, collaborating, and identifying patterns will be able to make faster and more effective decisions. In fact a new class of workers, often referred to as 'data scientists' or 'knowledge workers', is emerging to conduct exactly this type of Big Data analytical activity for both public and private organizations. However, Big Data also presents significant challenges related to the management, integration, updating, and visualization of information at this scale. Due to these challenges, computational tools that enable users to manipulate, visualize, and share in the analysis of data are currently, and will become evermore, important for enabling the derivation of knowledge and insight from Big Data. In particular, software tools that allow users to share and collaborate in the derivation of high-level knowledge and insight from the vast sea of data available are critical. The need for such tools is particularly pressing in the biomedical research and development space where the integration and understanding of relationships between information from multiple disparate data sources is crucial for accelerating hypothesis generation and testing in the pursuit for disease treatments.
19.2 Semantic technologies
A range of software tools have been developed over the past couple of decades to address the growth of Big Data across many sectors. Highly prevalent and successful text-based search tools such as Google [8], Endeca [9], FAST [10], and Lucene/Solr [11] (and discussed in Chapter 14 by Brown and Holbrook) enable users to conduct key word searches against unstructured text repositories with the results of these searches being lists of matching text documents. Although this sort of search paradigm and the tools that enable it are highly valuable, these tools have a number of attributes that are not optimally suited to Big Data scenarios. These attributes include:
![]() As the amount of data grows, the results derived from text-based search systems can themselves be so numerous that the user is left needing to manually sift through results to find pertinent information.
As the amount of data grows, the results derived from text-based search systems can themselves be so numerous that the user is left needing to manually sift through results to find pertinent information.
![]() Text-based search systems do not provide a means with which to understand the context and interconnections between resulting text. This is because results are generated through key word matching, not a formalized representation of the things (i.e. entities) that a user may search for, their properties and the associations between them.
Text-based search systems do not provide a means with which to understand the context and interconnections between resulting text. This is because results are generated through key word matching, not a formalized representation of the things (i.e. entities) that a user may search for, their properties and the associations between them.
![]() Certain disciplines, such as biomedical research and development, are confronted with the challenge that any given entity of interest may have many names, synonyms, and symbols associated with it. Because text-based search systems are based on key word matching, users can miss important search results if they do not search for each of the various names, synonyms, and symbols used to represent a given entity of interest.
Certain disciplines, such as biomedical research and development, are confronted with the challenge that any given entity of interest may have many names, synonyms, and symbols associated with it. Because text-based search systems are based on key word matching, users can miss important search results if they do not search for each of the various names, synonyms, and symbols used to represent a given entity of interest.
As an example of the difficulties associated with text-based search systems for biomedical research, let us imagine the scenario that a user wants to search for all of the information available for the gene ABL1. First, ABL1 has many names and symbols such as ABL1, c-abl, and V-abl Abelson murine leukemia viral oncogene homolog 1. Second, ABL1 can be both a gene and a protein (the translation of the gene). Third, ABL1 is a gene found in many species including human, mouse, rat, and primate. So, our hypothetical user would start in a text-based search system by typing ABL1 and they would receive a large set of document links as a result. That search would only be run across text documents (i.e. unstructured data sources) and thus the user would not be provided with any information about the gene from structured databases or other data sources. Additionally, how does the user know that they have found all documents relevant to ABL1 and all of its synonyms, symbols, and identifiers instead of only those that mention ABL1? Further, how do they refine their search to results matching the gene versus protein version of ABL1 and also the right species? Confronted with this set of challenges, it is often the case that the user simply does not address any of these issues and instead just accepts the results provided and attempts to sift through large quantities of results in order to identify those documents most relevant to their interests. In addition to the difficulties associated with finding all pertinent information for a given entity, it is often the case that the user is not only interested in the single entity but also for other entities with which it might be associated. For example, for a given gene product, like ABL1 protein, there may be compounds that are selective for it, diseases in which it is indicated, and clinical trials that might target it. All of the associations for a user's searched entity (e.g. ABL1) simply are not provided by text-based search systems, and instead the user once again must read through all resulting documents in order to try to glean associations for their search term. By forcing a user to manually read through large sets of documentation, critical properties of entities and knowledge about associations between entities that might be highly valuable to their research can be easily missed. To sum up, text-based search systems, although valuable tools, lack a range of attributes that would be useful in scientific and related disciplines in which a user is interested in searching for and analyzing structured data representations of entities and their associations.
Novel technologies have been developed over the course of the past two decades that make it possible to address many of the downsides of text-based search systems outlined above. These novel technologies adopt a 'semantic' approach to the representation of entity properties and their associations. With these technologies it is now possible to design and build next-generation systems for scientific research, which can handle large-volumes of data (Big Data), provide comprehensive search results to a user query no matter what names might be used to query for a given entity, and also automatically and dynamically show the user all of the other entities known to be associated with their queried entity.
19.3 Semantic technologies overview
The foundational set of 'semantic' technologies were originally proposed in 2001, by Tim Berners-Lee and colleagues in a Scientific American article entitled, 'The Semantic Web' [12]. The idea of the Semantic Web was to make information about entities and their associations with other entities accessible for computation. A range of flexible, open standard semantic technologies have been developed over the last decade under the leadership of the World Wide Web Consortium (W3C). These technologies enable novel forms of facile integration of large quantities of disparate data, representation of entities and entity associations in a computable form. Combined with comprehensive querying and inferencing capabilities (the ability to compute novel relationships 'on-the-fly'), semantic technologies promise collaborative knowledge building through joint creation of computable data. Furthermore, the foundational components of semantic technologies adhere to open, free standards that make them an excellent framework on which to build next-generation 'Big Data' search and analysis systems.
Semantic technologies utilize a data formatting standard called resource description framework or 'RDF' [13]. RDF defines a very simple but highly flexible data format called 'triples.' A triple is made up of a subject, a predicate and an object. Here is an example:
Most human languages have evolved to require even the most basic sentence to include a subject and a predicate (grammatically speaking, a predicate is inclusive of the verb and object). To compose a sentence is to assemble a snippet of meaning in a manner that can be readily interpreted by the recipient. An RDF triple is the codification of this requirement in a format suitable for computation. The triple is, to a degree, a basic sentence that can be intelligibly parsed by the computer. In short, RDF provides a standard for assigning meaning to data and representing how it interconnects, thus using semantics to define meaning in computable form. This simple but powerful design allows for computable forms of not only entity information but also the associations between entities.
In addition to the advantages provided by the flexibility of the RDF format, technologies for use with RDF also offer some compelling advantages. For example, RDF storage systems, commonly referred to as 'triple stores,' require little design. In comparison, the data models in relational databases can comprise dozens of tables with complex relationships between both the columns of a single table and between the tables. Diagrams of these tables, their columns, and interconnections are called a relational schema. The complexity of relational schemas can mean that administrators intent on updating a relational data model may need days or weeks to fully comprehend what will happen when values are modified in any given record. Furthermore, attempting to integrate multiple schemas from multiple databases can take months of work. An RDF triple store on the other hand comprises nothing more than a series of triples. This means that the type and properties for all of the things represented in the triple store are codified in one format with a standardized structure. As a result, integration across RDF triple stores, or the inclusion of new triples in a triple store, is as easy as combining sets of triples. Simply put, relational database schemas are fundamentally a non-standardized data format because each schema is different. In comparison, RDF is fundamentally a standardized data format thereby enabling greater data integration flexibility and interoperability.
RDF triple stores utilize a specialized query language called SPARQL [14] that is similar to the query language for relational databases, SQL. Despite the similarity between SPARQL and SQL, triple stores are easier to query because their contents are not partitioned into tables as is usually the case with relational data models. Furthermore, triple stores that are version-compliant can be swapped in and out relatively easily. The same cannot be said of relational database technologies where schemas usually require careful modification when being migrated or interconnected with a different database.
RDF triple stores are also comparatively fast thanks to the simple and fixed triples data format that allows for optimized indexes. This coupled with the inherent scalability available in many triple stores means programs can be written that provide views derived in real time from data present in billions of triples. Many RDF triple stores offer linear performance gains by scaling across many computers without the complex setup that accompanies most traditional relational database clusters. Triple stores can also readily scale across machines, as they do not have to support the complex data relationships associated with SQL-type databases. This characteristic relieves the triple store of the heavy overhead associated with locking and transactions.
All of the properties of semantic technologies including both the flexible, extensible formatting of data as RDF triples and the handling of data with the types of technologies outlined above, make these technologies an excellent foundation for next-generation search and analysis systems for scientific research and related Big Data applications. Of course, in addition to a data format and technologies to handle data, a biomedical search and analysis system would need actual data to be a valuable tool in a researcher's toolbox. Thankfully, there exists an excellent set of publically available biomedical data referred to as the Linking Open Drug Data set for use as a foundation.
19.3.1 Linking Open Drug Data
In the biomedical research and development (R&D) sector, flexible data integration is essential for the potential identification of connections across entity domains (e.g. compound to targets, targets to indications, pathways to indications). However, the vast majority of data currently utilized in biomedical R&D settings is not integrated in ways that make it possible for researchers to intuitively navigate, analyze, and visualize these types of interconnections. Collection, curation, and interlinking public biomedical data is an essential step toward making the sea of Big Data more readily accessible to the biomedical research community. To this end, a task force within the W3C Health Care and Life Sciences Interest Group (HCLSIG) [15] has developed the Linking Open Drug Data (LODD) set [16]. Standards for the representation of data within the LODD set have been defined and all of the data have been made available in the RDF format. The LODD is an excellent resource for the biomedical research and development community because it provides the basis for the interconnection of valuable biological, chemical and other relevant content in a standard format. This standardization of formatting and nomenclature is the critical first step toward an integrated, computable network of biomedically relevant data. It also provides the foundation for the formatting and integration of further data sets over time.
19.4 The design and features of TripleMap
Based on experience working with Big Data and in the biomedical research and development sector, we set out over three years ago to design and develop a next-generation semantic search and analysis system, which we named TripleMap. Users are welcome to register and login to the free web-based version of TripleMap at www.triplemap.com in order to test out its various features. The public instance of TripleMap is completely functional, lacking only the administrator features found in Enterprise deployments within an organization. Below, we outline TripleMap's features and then provide additional details regarding three aspects of the system, the Generated Entity Master (GEM) Semantic Data Core, the TripleMap semantic search interface, and collaboration through knowledge map creation and sharing.
TripleMap has a number of features that distinguish it from more traditional enterprise text-based search systems. In particular, it enables collaborative knowledge sharing and makes it possible for organizations to easily build an instance behind their firewall for their internal data. These features are outlined here.
![]() Text search equivalent to the current best of breed systems: searching in the TripleMap system is comprehensive including the entire contents of all documents in the system. It also provides relevancy scoring in a manner similar to text search engines such as Google, FAST, and Endeca.
Text search equivalent to the current best of breed systems: searching in the TripleMap system is comprehensive including the entire contents of all documents in the system. It also provides relevancy scoring in a manner similar to text search engines such as Google, FAST, and Endeca.
![]() Next-generation semantic search: as a next-generation semantic search platform, TripleMap goes beyond the capabilities of text search engines by giving users the ability to search not only for text in documents but also for entities, their associations, and their meta-data.
Next-generation semantic search: as a next-generation semantic search platform, TripleMap goes beyond the capabilities of text search engines by giving users the ability to search not only for text in documents but also for entities, their associations, and their meta-data.
![]() Display and navigation of entity to entity associations: upon finding a given entity, the user is able to easily identify other entities which are associated with their original input. Furthermore, the user is able to navigate through associations and identify novel associations as they move through information space.
Display and navigation of entity to entity associations: upon finding a given entity, the user is able to easily identify other entities which are associated with their original input. Furthermore, the user is able to navigate through associations and identify novel associations as they move through information space.
![]() Automated extraction of entity properties, labels, and associations: TripleMap is able to automatically derive the properties, labels, and associations for entities from the data it is given. Instead of having to laboriously create all of the data connections the system is going to use, administrators are able to provide the system with data sources and some basic information about those data sources and the system automatically derives and interconnects all of the data that it is given. The system is able to conduct this process of automated property, label, and association derivation continuously as novel data become available in all of the originating sources it is monitoring.
Automated extraction of entity properties, labels, and associations: TripleMap is able to automatically derive the properties, labels, and associations for entities from the data it is given. Instead of having to laboriously create all of the data connections the system is going to use, administrators are able to provide the system with data sources and some basic information about those data sources and the system automatically derives and interconnects all of the data that it is given. The system is able to conduct this process of automated property, label, and association derivation continuously as novel data become available in all of the originating sources it is monitoring.
![]() Derive data from numerous sources: TripleMap is able to derive data from numerous sources including static RDF files, dynamic remote SPARQL endpoints, relational databases, XML files, tab-delimited text files, Sharepoint TeamSites, RSS feeds, Documentum repositories and networked file system locations.
Derive data from numerous sources: TripleMap is able to derive data from numerous sources including static RDF files, dynamic remote SPARQL endpoints, relational databases, XML files, tab-delimited text files, Sharepoint TeamSites, RSS feeds, Documentum repositories and networked file system locations.
![]() Entity sets as persistent associative models or 'maps': TripleMap users are able to create maps of entities as they conduct searches. These maps are structured data representations of everything known in the system including entity properties, entity-entity associations and relevant documents. These maps can be saved and thus persisted across usage sessions in the system.
Entity sets as persistent associative models or 'maps': TripleMap users are able to create maps of entities as they conduct searches. These maps are structured data representations of everything known in the system including entity properties, entity-entity associations and relevant documents. These maps can be saved and thus persisted across usage sessions in the system.
![]() Maps as composite search strings against all information streams: any map a user creates is automatically used by the system as a composite search against all information streams that the system monitors. The system constantly scans these information streams (e.g. documents, RSS feeds, wikis, patent literature) for mentions of any of the entities in a user's saved maps and alerts the user if novel information is detected. Because the system is able to handle synonyms, symbols, and many names for any given entity, all mentions of a given entity, no matter what the name, are tied back to the 'master entity' representation of that entity.
Maps as composite search strings against all information streams: any map a user creates is automatically used by the system as a composite search against all information streams that the system monitors. The system constantly scans these information streams (e.g. documents, RSS feeds, wikis, patent literature) for mentions of any of the entities in a user's saved maps and alerts the user if novel information is detected. Because the system is able to handle synonyms, symbols, and many names for any given entity, all mentions of a given entity, no matter what the name, are tied back to the 'master entity' representation of that entity.
![]() Automated entity alerting: the user is alerted to the appearance of novel information relating to any of the entities in any of their maps. Alerting is available through email, RSS feeds and by viewing maps in the system.
Automated entity alerting: the user is alerted to the appearance of novel information relating to any of the entities in any of their maps. Alerting is available through email, RSS feeds and by viewing maps in the system.
![]() One data model, multiple views: TripleMap provides multiple views of the map data and each view allows for an alternative visualization of the interconnection of entities in a map.
One data model, multiple views: TripleMap provides multiple views of the map data and each view allows for an alternative visualization of the interconnection of entities in a map.
![]() Set sharing and searching: users are able to both share the maps that they create with colleagues and easily search through all available maps in order to identify colleagues who are currently working on similar maps or have created and saved similar maps in the past.
Set sharing and searching: users are able to both share the maps that they create with colleagues and easily search through all available maps in order to identify colleagues who are currently working on similar maps or have created and saved similar maps in the past.
![]() Collaborative knowledge building through de novo user-defined associations: users are able to create novel associations between entities in the system and set the visibility of those de novo associations to private, group-based, or public. The appropriate path back to the originating creator of an association is visible to all users to allow for validation of user-generated associations.
Collaborative knowledge building through de novo user-defined associations: users are able to create novel associations between entities in the system and set the visibility of those de novo associations to private, group-based, or public. The appropriate path back to the originating creator of an association is visible to all users to allow for validation of user-generated associations.
![]() Web-based application: the system employs novel software technologies that allow for the provision of rich user experiences through a web-based application. Web-based applications provide a number of key advantages including ease of administration, deployment, and updating.
Web-based application: the system employs novel software technologies that allow for the provision of rich user experiences through a web-based application. Web-based applications provide a number of key advantages including ease of administration, deployment, and updating.
![]() Data-driven platform: the system is data-driven and flexible enough so that any given instance of the system can be tailored to the specific interests and needs of the administrator.
Data-driven platform: the system is data-driven and flexible enough so that any given instance of the system can be tailored to the specific interests and needs of the administrator.
![]() Massively scalable: TripleMap scales both in terms of its ability to support large-scale data sets and its ability to support large numbers of concurrent users. A range of high-performance technologies such as computer clustering and high-performance indexing are employed to enable this.
Massively scalable: TripleMap scales both in terms of its ability to support large-scale data sets and its ability to support large numbers of concurrent users. A range of high-performance technologies such as computer clustering and high-performance indexing are employed to enable this.
![]() Advanced association analytics: the system allows users to perform advanced analytics against the full underlying data network provided by all entities and all of the associations between entities available in the system. For example, users are able to infer high-order connections between any two entities in the system.
Advanced association analytics: the system allows users to perform advanced analytics against the full underlying data network provided by all entities and all of the associations between entities available in the system. For example, users are able to infer high-order connections between any two entities in the system.
19.5 TripleMap Generated Entity Master ('GEM') semantic data core
At the heart of the TripleMap system is a large-scale, high-performance data core referred to as the TripleMap Generated Entity Master ('GEM') (the TripleMap architecture with the central GEM data core is shown in Figure 19.1 ). The data GEM contains the entire master data network available within the TripleMap system and can be continually updated and enhanced by TripleMap administrators as they identify more data sources for integration. The GEM controller is used to aggregate and integrate data from a variety of sources (e.g. RDF, flat file, relational databases, Sharepoint, RSS feeds, patent literature). TripleMap users interact with this data core by running semantic searches and then storing results as sets, or maps of entities and the associations between entities.
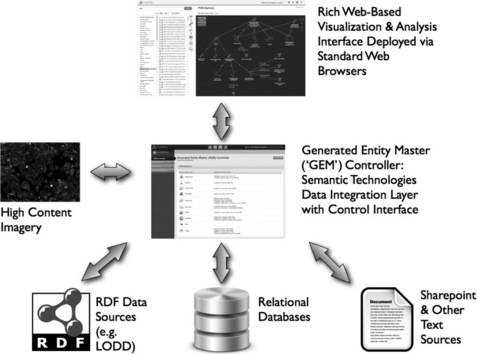
Figure 19.1 The TripleMap architecture. The GEM is the central semantic data core of any given TripleMap instance
These sets are sub-networks of data derived from the full master data network based on the user's interests. Maps can be shared and dynamically updated thereby enabling collaborative knowledge sharing.
TripleMap proprietary GEM technology is based on a combination of systems for integrating data across data sources and indexing those data for high-performance search results, along with the use of a standards-compliant triple store for storage of the originating data. Any of a number of triple stores including bigdata®, Sesame, Allegrograph or 4Store can be used with TripleMap. In current installations, the TripleMap architecture in association with a standards-compliant triple store is capable of handling > 1 billion triples with millisecond time performance for the return of search results.
In building a new instance of TripleMap, an administrator starts by importing multiple data sources for integration into the GEM controller. Once imported, the administrator determines which entities in the originating data sources should be combined and represented in TripleMap. The administrator then creates 'master entities' and through a simple application interface ties entities from each data source through to the appropriate 'master entities'. Once this data modeling and linkage process is complete, the administrator clicks the 'Generate GEM' button and TripleMap initiates a completely automated, comprehensive process referred to as 'data stitching'. During data stitching, entities from the originating sources are integrated based on a set of factors including labels and associative predicates. TripleMap is able to automatically derive and generate all of the integrated representations of entity properties, the associations between entities and the full set of names, synonyms, and symbols for each entity. This process of data import, integration, and stitching can be conducted at any time for a TripleMap instance, and can be run whenever new data become available, thereby allowing administrators to continuously and iteratively update the master data network for users. This full process of GEM creation and administration is performed routinely for the public instance of TripleMap at www.triplemap.com.
One of the key advantages of the TripleMap GEM data core is that it provides a mechanism through which administrators can set up and maintain a large-scale, 'master' network of integrated data (Figure 19.2). TripleMap users search and collaborate using this master data network. This concept of a centralized data network differs from applications where users upload data on a use-by-use basis in order to analyze and interact with the data. Instead TripleMap is a comprehensive, continuously updating system that provides a means of searching and interacting with a large-scale, pervasive network of entity-relevant data and associations between entities.
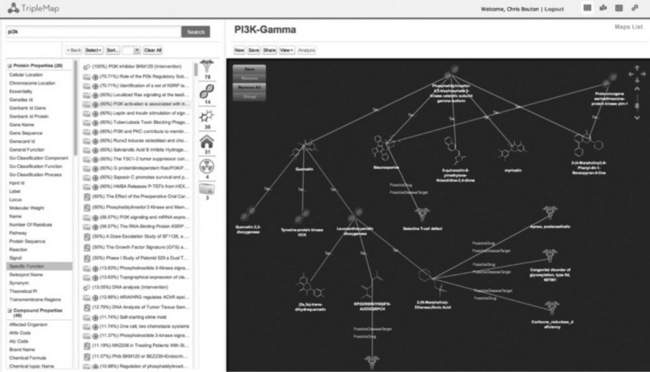
Figure 19.2 Entities and their associations comprise the GEM data network. The TripleMap GEM data network is generated through the representation of entities (e.g. proteins, genes, diseases, assays) and the associations between those entities (e.g. activation, inhibition, associations). Each knowledge map created by a user is a subnetwork of the master entity data network created within the GEM semantic data core
19.6 TripleMap semantic search interface
TripleMap is designed as a semantic search interface with the added capability for users to store and share sets, or maps, of structured search results. The sets of entities that are stored by users have both meta-data properties and entity to entity associations and hence can also be viewed as maps of those entities and the associations between them (Figure 19.3). The left-hand panel of the web application contains a semantic search interface including entity search, faceted filtering and quick link capabilities. Users can drag and drop entities from their search results into the knowledge map view in the right-hand panel. This interplay between searching and collecting sets of entities in knowledge maps is at the heart of the TripleMap usage paradigm. With one or more entities selected, further connections are shown as icons and counts to the right of the search panel. Users can see a list of all of their maps with the Maps List view and also a printout of all properties of each entity as a 'Semantic Knowledge Review' in the right-hand panel. Using the Maps List or Map view interfaces, users can securely share maps with each other via email invites and receive alerts (either email or RSS) to new information that is published or added to the GEM data core about interesting entities.
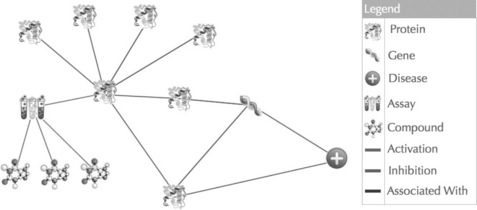
Figure 19.3 TripleMap web application with knowledge maps. The TripleMap web application interface is shown. Knowledge maps are created from search results and then saved and shared by users of the TripleMap system. Maps are used by the system as automatic scans against all unstructured (e.g. documents, journal articles, patent text, RSS feeds) sources available to the system such that novel information about anything in any map is highlighted and the user is alerted (via email or RSS) once it is detected
19.7 TripleMap collaborative, dynamic knowledge maps
The rapid growth of the internet has fostered the development of novel 'social media' technologies. These technologies enable large communities of individuals to share information, communicate and interact. The rapid adoption of these systems by user communities speaks to their value for human interaction. As successful as these systems are however, many have very limited semantic capabilities. Twitter [17], for example, allows for nothing more than the sharing and forwarding of small snippets of text (140 characters to be exact) with the 'hashtag' ('#') serving as the only meta-data feature. Facebook [18], another highly successful social system, allows for the simple sharing of personal information and photos. YouTube [19] allows for the sharing of video and LinkedIn [20] allows for the sharing of one's professional information. These are all fairly simple systems and yet they have become massively pervasive, impactful, and successful. As discussed in the chapters by Alquier (Chapter 16), Wild (Chapter 18) and Harland et al. (Chapter 17), we share the vision that as semantic technologies establish themselves, the next generation of collaboration tools will evolve through the more sophisticated capabilities enabled by computable knowledge representation. With that in mind we have designed TripleMap so that it provides an extension of the collaborative capabilities of platforms such as wikis, Twitter, and social networks by utilizing semantic technologies to enable users to share and collaborate around the creation of structured data representations or maps of entities and the associations between entities. The maps in TripleMap are structured data representations of what is known about the entities in a given domain space (e.g. drugs, diseases, targets, clinical trials, documents, people, organizations, the meta-data for those entities, and the associations between them). Maps are created as users search for and identify the entities that are of interest to them. Furthermore, we refer to the maps that are created by users as 'dynamic knowledge maps' (Figure 19.3 ) because once created by a user any given map is continuously and automatically updated with the latest information being published from structured (e.g. databases) and unstructured (e.g. document) sources.
Knowledge maps promote collaboration because once built they can be shared with colleagues in private groups or can be shared publicly with the entire TripleMap community. Each knowledge map provides a 'bird's eye view' of the things in which a user is most interested (Figure 19.3). Users are able to save and share the knowledge maps that they create. The creation and saving of maps is valuable for several reasons. First, in the act of creating maps, the associations, including unexpected associations, between things are discovered because the system prompts the user with all associations between entities that are stored in the master data network of the GEM data core. Second, maps can be shared with others, thereby allowing users to share structured data representations of the information in which they are interested with their colleagues in groups or more broadly with the wider research community. Third, users can search through all available maps to identify other users working on maps similar to theirs. Finally, maps can be used for auto-searching against all new information being scanned by the system and alerting a user if novel information is available for one or more of the things in which they are interested.
Knowledge building through the sharing of knowledge maps is similar to what is seen in wiki communities where users collaborate around the development of pages of text that describe various topics. The difference between shared map building and shared wiki page building is in the structuring of the data which users are pulling from to create maps in the first place. TripleMap knowledge maps are effectively subnetworks of structured, interlinked data describing things and the associations between them. These subnetworks are portions of the entire master data network integrated and continuously updated in the TripleMap GEM. In the wiki scenario there is less structuring of the content generated and it is more difficult to identify interrelationships between the things mentioned in pages. Despite the differences between TripleMap and wikis, there are valuable ways in which these two types of systems can be linked together in order to provide a more comprehensive collaborative platform for biomedical research. For example, integration of a system like the Targetpedia (see Harland et al., Chapter 17) with TripleMap allows users to interact with information about targets both as a text-based wiki page and then also link out to knowledge maps related to each target within one integrated system.
19.8 Comparison and integration with third-party systems
When used in an enterprise context, TripleMap is not redundant with general-purpose document search systems such as FAST or Google (both are discussed above). Instead TripleMap provides a framework and application for the semantic search that enhances the utility of enterprise document search systems and can be used in conjunction with enterprise search frameworks such as Google or the FAST search engine.
Additionally, TripleMap is not redundant with applications focused on quantitative data analytics such as Spotfire [21] or Tableau [22]. Instead TripleMap provides an environment in which users can conduct semantic searches across a large-scale shared master network of data in order to identify and build 'bird's eye view' representations of what is known in a given information space. Software bridges to specialized systems such as Spotfire or Tableau through available third-party API bridges are being developed so that users can conduct further quantitative data analysis on entities identified and monitored in TripleMap.
19.9 Conclusions
TripleMap is a next-generation semantic search, knowledge discovery and collaboration platform. We have provided an instance of TripleMap based on data from the Linking Open Drug Data sets for free to the biomedical research community at www.triplemap.com. Organizations can also bring TripleMap internally for use with proprietary data using the TripleMap Enterprise platform. This has indeed been the case with a number of commercial life science companies who are using the technology to integrate their internal data and documents alongside public content. TripleMap is built on and extends open standards and semantic technologies developed primarily by the W3C. These open standards are critically important for the uptake of solutions based on these standards and the next-generation data-handling capabilities that they enable. We believe that the future holds tremendous promise for the derivation of insights from the vast troves of Big Data available to humanity. Furthermore, we believe that the design, development, and deployment of software systems that enable this derivation of insight from Big Data sources is crucial. Our goal in making the www.triplemap.com instance of TripleMap available for free to the biomedical research community is to foster collaborative interaction and discovery in the pursuit of the development of fundamental, ground-breaking treatments for human diseases.
19.10 References
[2] http://www.w3.org/wiki/HCLSIG/LODD.
[3] Gantz, B.J., Reinsel, D. Extracting Value from Chaos State of the Universe? An Executive Summary. 2011; 1–12. [(June)].
[4] Hal, R., Varian, PL., How Much Information?, 2003. Retrieved from http://www2.sims.berkeley.edu/research/projects/how-much-info-2003/
[5] Hilbert, M., Lopez, P. The world's technological capacity to store, communicate, and compute information. Science (New York, N.Y.). 2011; 332(6025):60–65. [10.112/science.1200970].
[6] Manyika, J., Chui, M., Brown, B., Bughin, J., Dobbs, R., Roxburgh, C., Big data: The next frontier for innovation, competition, and productivity, 2011. [(May).].
[7] Lander, E.S., Linton, L.M., Birren, B., et al, Initial sequencing and analysis of the human genome. Nature. 2001;409(6822):860–921, doi: 10.1038/35057062.
[10] http://www.microsoft.com/enterprisesearch/en/us/fast-customer.aspx.
[11] http://lucene.apache.org/java/docs/index.html.
[12] Berners-Lee, T., Hendler, J. Lassila O. Scientific American: The Semantic Web; 2001.
[13] http://www.w3.org/RDF/.
[14] http://www.w3.org/TR/rdf-sparql-query/.
[15] http://www.w3.org/wiki/HCLSIG.
[16] Samwald, M., Jentzsch, A., Bouton, C., et al, Linking Open Drug Data for pharmaceutical research and development. Journal of Cheminformatics. 2011;3(1):19, doi: 10.1186/1758-2946-3-19. [Chemistry Central Ltd.].
[17] http://www.twitter.com.
[18] http://www.facebook.com.
[19] http://www.youtube.com.
[20] http://www.linkedin.com.
