Setting up an ’omics platform in a small biotech
Abstract:
Our current ’omics platform is the product of several years of evolution responding to new technologies as they have emerged and also increased internal demand for tools that the platform provides. The approaches taken to select software are outlined as well as any problems encountered and the solutions that have worked for us. In conjunction lessons learnt along the way are detailed. Although sometimes ignored, the range of skills required to use and maintain the platform are described.
10.1 Introduction
Founded in 1995 by the pioneer of Southern blotting and microarray technologies, Professor Sir Edwin Southern, Oxford Gene Technology (OGT) is focused on providing innovative clinical genetics and diagnostic solutions to advance molecular medicine. OGT is based in Yarnton, Oxfordshire and is a rapidly growing company with state-of-the-art facilities and over 60 employees. The key focus areas of OGT are:
![]() tailored microarray [1] and sequencing services enabling high-quality, high-throughput genomic studies;
tailored microarray [1] and sequencing services enabling high-quality, high-throughput genomic studies;
![]() cytogenetics products and services delivering the complete oligonucleotide array solution for cytogenetics;
cytogenetics products and services delivering the complete oligonucleotide array solution for cytogenetics;
![]() delivering tailored biomarker discovery solutions to identify and validate the best biomarkers for all diagnostic applications;
delivering tailored biomarker discovery solutions to identify and validate the best biomarkers for all diagnostic applications;
![]() licensing fundamental microarrays and other patents;
licensing fundamental microarrays and other patents;
![]() developing new diagnostic techniques with potential for in-house commercialisation or partnering with diagnostic and pharmaceutical organisations;
developing new diagnostic techniques with potential for in-house commercialisation or partnering with diagnostic and pharmaceutical organisations;
![]() delivering a comprehensive sequencing service using next-generation sequencing technologies.
delivering a comprehensive sequencing service using next-generation sequencing technologies.
Here, I will describe the approach that OGT has taken in the development of our in-house, ’omics platform. This covers the initial stages of setting up a commercial platform to service customer microarrays, through to developing our own microarray products, running high-throughput projects to recently setting up a pipeline to assemble and annotate data from next-generation sequencers. Each of these has required a range of software applications. Whenever OGT requires new software, a standard approach of evaluating all possible options is followed, be they FLOSS, commercial or even commissioned; weighing up the advantages and disadvantages of each solution. Obviously an important consideration is cost, and where there are comparable products then this becomes an important part of any discussion to decide which products to use. However, cost is just one of a series of important factors such as ease of use, quality of output and the presence/absence of specific skills within the group. Furthermore, any likely impact on infrastructure must also be borne in mind before a decision is made. In reality, more often than not a FLOSS alternative will be selected, indicating just how much high-quality FLOSS software is now available.
10.2 General changes over time
The computational biology group at OGT has evolved hugely over this time. In 2003, OGT was a small biotechnology company with a staff numbering in the low 20s, servicing mostly small academic projects for customers within the UK. Since then, the company has grown to more than 70 full-time employees and has customers from both industry and academia based around the globe. Concomitantly, the typical value of projects has greatly increased from the low thousands of pounds involving a handful of arrays, to hundreds of thousands based on thousands of microarrays. To cope with this growth, our internal procedures and processes have had to develop and evolve.
Indeed, the consequences of the improvements in microarray technology have been the increased demands placed on the IT infrastructure that supports it. For example, in 2003 the densest microarray that OGT produced contained 22 500 oligonucleotide probes. Following hybridisation and processing, this generated a data file of about 12 MB. Since then, DNA spotting has improved such that more features can be printed, and the resolution of scanners has similarly increased so these features can be smaller. This frees up real estate on the slide allowing for increases in feature number. At the time of writing, the latest arrays from our microarray provider, Agilent, have 1 000 000 features and generate a data file around 1.4 GB. This dramatic increase impacts across the IT system; files take longer to move around the network, backups take longer to run and files are now too big to deliver by email or via traditional methods such as FTP. Furthermore, we have recently expanded our offerings to include next-generation sequencing (NGS). This has necessitated a large investment in hardware to provide the processing power to deal with the terabytes of data generated. The chapters by Thornber (Chapter 22) and MacLean and Burrell (Chapter 11) provide good background on NGS data for those unfamiliar with this topic.
10.3 The hardware solution
Within the company there is a requirement for our hardware system to integrate the needs of the computational biology team, who use both Windows and Linux, with those outside the group, who work purely within a Windows environment. For example, the PCs in the labs used to process the microarrays must be able to write to a location that can be accessed by the Linux-based applications used to process the data. The approach taken at the start of OGT, and still in place today, was to keep all data on disk arrays controlled by a Linux server. These directly attached storage (DAS) arrays mounted on the Linux platform are then made available to all users over the company network. This is accomplished using the Linux utility Samba [2]. Samba comes with a great deal of supporting information to allow users to understand this powerful system. Essentially, there are folders defined within Samba to which Windows users can map a drive. The appearance of these drives is of a normal Windows folder, but under the bonnet the information is being made available across the network from a Linux server. This integration of Windows and Linux is reinforced by the fact that all authentications are handled by Windows Active Directory. It is the Active Directory that controls all aspects of user permissions, irrespective of the platform being used, making it much easier to manage the system. The only exceptions to this are root access to the servers, which is independent and will authenticate as usual via the standard/etc/ passwd mechanism.
Originally, the first hardware purchased for the OGT ’omics platform was a complete system comprising a Beowulf compute cluster of a master server and four nodes. The master server handled all processes and farmed out jobs to the nodes on the cluster. All data was stored on a 1 TB DAS disk array and there was tape library to handle backups. This is shown in Figure 10.1.
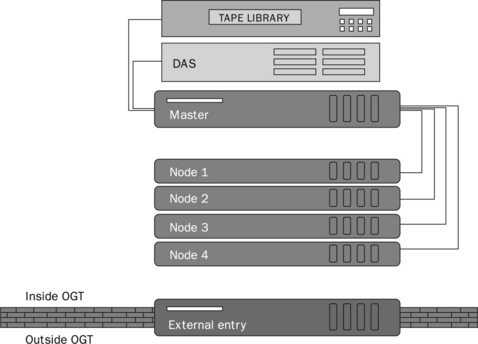
Figure 10.1 Overview of the IT system showing the Beowulf compute cluster comprising a master server that passes out jobs to the four nodes. The master server also provides access to the DAS disk array and performs data backups using a tape library
The advantage of buying a pre-configured system was that it was ready to go on arrival. It reduced the time required to get up and running and make the ’omics platform operational. Also, although some of the group had advanced Linux skills, these were short of the level needed to act as a true system administrator and setup a system from scratch. Following installation by our supplier, all day-to-day maintenance could be handled by a nominated member of the computational team who had access to an external consultant if required. The disadvantage of buying a pre-configured system became apparent when there was a problem after the warranty had expired. The master server developed a problem in passing jobs to the compute nodes. The supplier would no longer support the system and our Linux consultants could not resolve the issue because they did not understand how the cluster was meant to be working.
In the end, the situation was resolved by the purchase of a new server. With this server, and all subsequent servers, we have taken the approach of buying them without any operating system (OS) installed. We are therefore able to load the OS of our choice and configure the server to our own individual requirements. This can delay the deployment but it does mean when there are problems, and there have been some, we are much better able to resolve the issue.
Lesson: Understand your own hardware or know somebody who does.
This highlighted two other important points. First, the design of our system as it was with a single server meant there was no built-in redundancy. When the server went down, the disk arrays went down. Consequently, none of the Windows folders mapped to the disk array were available; this had an impact across the whole of the company. Second, it showed that IT hardware needs replacing in a timely manner even if it is working without fault. This can be a difficult sell to management as IT hardware is often viewed with the mantra ‘if it ain’t broke don’t fix it’; however, at OGT we take the approach that three years is the normal IT lifespan and, although some hardware is not replaced at this milestone, we ensure that all the hardware for our business critical systems is. This coupled with a warranty that provides next business day support, means that OGT has a suitable level of resource available in the event of there being any IT hardware issues.
Lesson: Take a modular approach and split functions between systems.
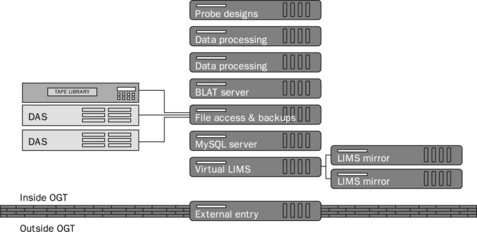
Subsequently, we have developed a more component-based approach of dividing operations between different servers so that even when one server goes down unexpectedly, other services are not affected. For example, there is a single server providing file access, another for running a BLAT server, another running our MySQL database, etc. This is illustrated in Figure 10.2.
10.4 Maintenance of the system
Responsibility for the day-to-day administration of the ’omics platform resides within the computational biology group. There is a nominated system administrator, but we also have a support contract with a specialist Linux consultancy who are able to provide in-depth knowledge when required. For example, when we buy a new server, they install the operating system and configure it for our needs. Also, they provide a monitoring service so that all our systems are constantly being assessed, and in the event of a problem being detected are alerted and able to resolve it. Further to this, they have been able to provide sensible and practical advice as we have expanded. From the OGT administrator’s point of view, this reduces time spent as a system administrator, freeing up time for other important functions in the group.
One early issue was that the Linux and Windows systems were maintained by separate people, with little communication flow in either direction. Given the overlap between these systems, this was not ideal. A recent development has been the formation of an IT steering committee comprising the Linux and Windows system administrators and some other IT literati from within OGT. With this has come regular fortnightly meetings at which any problems can be discussed, as well as any plans for the future discussed. Usually these meetings do not last very long, but they have significantly oiled the IT machinery within OGT.
Finally, with regard to being a system administrator, the following are some general rules of thumb I have found that hold true in our experience.
![]() Never do something you can’t undo.
Never do something you can’t undo.
![]() Always check the backups. Never assume they are working and make sure you can restore from them.
Always check the backups. Never assume they are working and make sure you can restore from them.
![]() Write down what you did, even if you know you will never forget it, you will. In our experience, DokuWiki [3] is an excellent documentation tool.
Write down what you did, even if you know you will never forget it, you will. In our experience, DokuWiki [3] is an excellent documentation tool.
![]() If you do something more than once, write a script to do it.
If you do something more than once, write a script to do it.
![]() Get to know your users before there is a problem. Then, when there is, they will know who you are and maybe have a little understanding.
Get to know your users before there is a problem. Then, when there is, they will know who you are and maybe have a little understanding.
![]() Remember you are performing a service for your users, you don’t own the system, you just get to play with it.
Remember you are performing a service for your users, you don’t own the system, you just get to play with it.
![]() Before running rm –rf *, always check your current directory; especially if you are logged in as a superuser.
Before running rm –rf *, always check your current directory; especially if you are logged in as a superuser.
![]() Never stop learning; there is always something you should know to make your job easier and your system more stable and secure.
Never stop learning; there is always something you should know to make your job easier and your system more stable and secure.
10.5 Backups
Given our commercial projects as well as our internal development projects, there is an absolute requirement to ensure that all necessary data are secured against the possibility of data loss. We settled on tapes as the medium for this, and our chosen backup strategy is to run a full backup of all data over the weekend and incremental backups Monday to Thursday. This covers all OGT’s current projects; once a project is completed it is archived to a separate tape volume and retained, but not included further in the weekly backups as it is no longer changing.
Initially, our pre-configured IT system was delivered with a commercial product installed, this was NetVault from BakBone [4]. As this was already installed and configured on the system, worked well and was easy to use, there was no need to change to an alternative. However, as the amount of our IT hardware expanded so did the licence requirements for running NetVault. As a result, we considered open source alternatives. The main candidates were Bacula [5] and Amanda [6] (the Advanced Maryland Automatic Disk Archiver). In the end, we chose to use Bacula primarily because it was the package with which our support was most familiar. Tools such as Bacula illustrate a common factor when considering FLOSS against commercial solutions, namely the level of GUI usability. However, in this case this was not such a factor as the console-based command-line controller offers a rich set of functionality. Although the learning curve is clearly steeper, this option means that administrators have no choice but to develop a better understanding of the tools capabilities as well as providing much finer control of the processes involved. Finally, the data format used by NetVault is proprietary and requires the NetVault software to access any previous archives. In contrast with this, Bacula provides a suite of tools that allow access to any archives without the requirement that Bacula is installed.
10.6 Keeping up-to-date
Taking this modular approach has helped significantly in managing the setup, as well as making the system much more robust and resistant to failure. Although the result is a more stable hardware platform, it has still been important to stay up-to-date with new technologies. For example, there is a great deal written about cloud computing and its impact on anything and everything (see Chapter 22 by Thornber, for example). Indeed, at a recent conference on ‘end-to-end’ infrastructure, most suppliers were offering products using virtualisation and cloud capabilities. This is something often discussed internally and something we feel does not offer any benefits significant enough to make it worth implementing at OGT at the moment. Of course, as this technology matures its capabilities will only increase and it will almost certainly have a future role to play at OGT. However, given the amount of data processed on a daily basis, the use of the cloud as a data repository is neither practical nor cost-effective at the moment. Where it may play a role for OGT is in providing a platform as a service; for example, if there was a short-term requirement for additional processing power that could not be met by our current systems. The ability of the cloud to provide such additional resources quickly and easily could prove very useful.
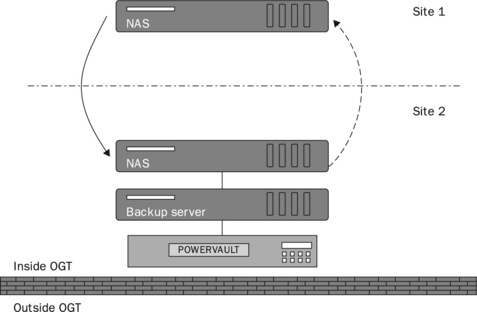
On a simpler level, there are alternatives to the DAS disk arrays currently in use. One of these is a network attached storage (NAS) device, as shown in Figure 10.3. As described previously, DAS disk arrays are used to provide the necessary data storage facility within OGT. However, some aspects of the system are being updated to include NAS devices as well. As the name suggests, a NAS device is a computer connected to the network providing storage, and, as such, performs a job similar to a DAS; but there are some significant differences. The key difference is that a NAS is a self-contained solution for serving files over a network, whereas a DAS is a set of disks made available by an existing server. The NAS units currently used already come with an embedded Linux operating system. Consequently, from the initial setup on the network through to its day-to-day management the NAS will require less administration than a DAS. Furthermore, all administration will be handled through GUI-driven management software. As such, it is close to a plug-and-play solution with all the necessary software pre-installed, for instance, for it to act as a file server, web server, FTP server or printer server. There is also the option of replication between NAS devices, which will further strengthen the platform. We employ one NAS box as the primary file server and use the replication software on the NAS to maintain a duplicate instance at another site. There are some disadvantages of using a NAS. As a pre-configured system it is generally not as customisable in terms of hardware software as a general-purpose server supplied with DAS. But, as all we require is a means to make files available over the network, the NAS is currently the best option.
10.7 Disaster recovery
One of the best definitions of disaster recovery is from Wikipedia [7] and is as follows:
Disaster recovery is the process, policies and procedures related to preparing for recovery or continuation of technology infrastructure critical to an organisation after a natural or human induced disaster.
‘What happens if the building is hit by a plane?’ is an often-quoted but fortunately rare scenario, but it remains the benchmark against which all recovery plans are measured. A perfect plan for disaster contingency will deliver zero data loss coupled with zero system downtime. Although this is always possible, such a level of protection will likely come at too high a price to be viable from a business perspective. At OGT, we have taken a flexible view of our needs and ensured that business critical systems have mechanisms in place to ensure they can be maintained. Other less important systems are given a lower level of support corresponding to their importance to the organisation.
There are different types of measure that can be employed to defend against disaster:
![]() preventive measures – these controls are aimed at preventing an event from occurring;
preventive measures – these controls are aimed at preventing an event from occurring;
![]() detective measures – these controls are aimed at detecting or discovering unwanted events;
detective measures – these controls are aimed at detecting or discovering unwanted events;
![]() corrective measures – these controls are aimed at correcting or restoring the system after a disaster or event.
corrective measures – these controls are aimed at correcting or restoring the system after a disaster or event.
These controls should be always documented and tested regularly. Documentation is particularly important; if the recovery plan does not accurately describe the process to restoring the system, the value of the entire plan is significantly lessened. Modularity again plays a key role here, allowing the plan to be tailored for different elements of the system, such as those described below.
10.7.1 Servers
Disk images of all servers are maintained such that they can be rebuilt on new hardware with a minimum of effort. The biggest delay that would be experienced would be in obtaining any new hardware.
10.7.2 Disk arrays
All disk arrays operate at RAID 6 level. In technical terms, this means this provides ‘block level striping with double distributed parity’; in practical terms, an array can lose up to two disks without any data loss. In the event of a disk failure (which has happened more than once), the faulty disk can be replaced with a new disk. The disk array automatically re-builds and the hardware is repaired.
Lesson: Buy spare disks when you buy the hardware to ensure compatibility or replacements.
A further enhancement is the use of hot spares. This is a disk physically installed in the array but in an inactive state. If a disk fails, the system automatically replaces the failed drive with a hot spare and re-builds the array. The advantage of this is that it reduces the recovery time. However, it can still take many hours for a disk to re-build; in our disk arrays 24 hours would normally be allowed for a disk to complete re-building. If there is a fault with the unit other than with a disk, then downtime will be longer. One of the advantages gained from having duplicate NAS boxes is that, in the event of a such a fault, we can switch over from the primary NAS to the secondary unit in order to maintain a functional system. With the new duplicate NAS boxes should there be a fault with the unit as opposed to a disk, then we can switch to the backup NAS box while the primary unit can be repaired.
10.7.3 Data
Besides ensuring that the hardware is stable, the data on the disks can also be secured using backups. It is worth considering what data absolutely must be saved and what could be lost. For example, prioritising the TIFF image files generated by the scanners from the processed microarray slides. All the data used in the analysis of the microarray are extracted from the image file and so can easily be regenerated as long as the original TIFF image file is available.
An additional measure we take is in backing up relational databases. The MySQL administration command mysqldump provides a useful mechanism to create an archive of all the information in the database. The resultant file contains everything required to recreate the database and restore any lost data. Similarly, all software development is managed with Subversion [8], a software versioning and revision control system. This not only provides a suitable backup, but also keeps track of all changes in a set of files, maintaining a record of transactions.
10.7.4 ’omics tools
Running an ’omics platform requires the use of a range of tools. A good place to start exploring the range currently available are the resources provided by the European Bioinformatics Institute (EBI). There are online versions of most tools, but local installations provide greater throughput for those run most frequently, also ensuring high-priority analyses are not subject to delays sometimes experienced on shared resources. Typically at OGT, there are local installations of major systems such as the Ensembl [9] Homo Sapiens genome database, providing immediate query responses as part of our processing pipeline. Pointing the same scripts to the same databases or web services held externally at the EBI would result in queries running much more slowly. When there are only a few queries this does not matter so much, but as the number of queries increases to hundreds and thousands then the impact is greater. A local installation is also critical from a security point of view, particularly where the data are confidential. Customers are always happier being told that all systems are internal, and there have been situations where this has been stipulated as an absolute necessity. Of course, there is the overhead of maintaining these installations, but in the long run this is a cost worth meeting. The following is a list of general FLOSS tools that are routinely used in our pipelines.
10.7.5 Bioinformatics tools
![]() BLAST [10] – almost certainly the most used ’omics application. The Basic Local Alignment Search Tool, or BLAST, allows comparison of biological sequence information, such as the amino acid sequences of different proteins or the nucleotide sequence of DNA. A BLAST search enables a researcher to compare a query sequence with a library or database of sequences, and identify library sequences that resemble the query sequence;
BLAST [10] – almost certainly the most used ’omics application. The Basic Local Alignment Search Tool, or BLAST, allows comparison of biological sequence information, such as the amino acid sequences of different proteins or the nucleotide sequence of DNA. A BLAST search enables a researcher to compare a query sequence with a library or database of sequences, and identify library sequences that resemble the query sequence;
![]() MSPcrunch [11] – this is a less well known but powerful tool that makes processing of large-scale BLAST analyses straightforward. Data can be easily parsed on a range of parameters. Also, there is a useful graphical view of the BLAST alignments that can be used in report generation applications;
MSPcrunch [11] – this is a less well known but powerful tool that makes processing of large-scale BLAST analyses straightforward. Data can be easily parsed on a range of parameters. Also, there is a useful graphical view of the BLAST alignments that can be used in report generation applications;
![]() EMBOSS [12] – this is a freely available suite of bioinformatics applications and libraries and is an acronym for European Molecular Biology Open Software Suite. The EMBOSS package contains around 150 programs covering a variety of applications such as sequence alignment, protein domain analysis and nucleotide sequence pattern analysis, for example, to locate repeats or CpG islands;
EMBOSS [12] – this is a freely available suite of bioinformatics applications and libraries and is an acronym for European Molecular Biology Open Software Suite. The EMBOSS package contains around 150 programs covering a variety of applications such as sequence alignment, protein domain analysis and nucleotide sequence pattern analysis, for example, to locate repeats or CpG islands;
![]() Databases – a relational database provides the backend to many applications, storing data from many sources. There are options available when choosing a suitable relation database system. These can be free and open source such as MySQL [13], PostgreSQL [14] and Firebird [15], or licence-driven products such as Oracle [16] and SQL Server [17]. We have chosen to use MySQL, primarily on the basis that it is the system we have most knowledge of so database administration is more straightforward. A strong secondary consideration was that it is free of charge. Also, the requirements for our database were relatively simple so pretty much any product would have been suitable.
Databases – a relational database provides the backend to many applications, storing data from many sources. There are options available when choosing a suitable relation database system. These can be free and open source such as MySQL [13], PostgreSQL [14] and Firebird [15], or licence-driven products such as Oracle [16] and SQL Server [17]. We have chosen to use MySQL, primarily on the basis that it is the system we have most knowledge of so database administration is more straightforward. A strong secondary consideration was that it is free of charge. Also, the requirements for our database were relatively simple so pretty much any product would have been suitable.
10.7.6 Next-generation sequencing
At OGT we have compiled a pipeline capable of processing data from next-generation sequencers. The pipeline takes in the raw data reads and runs a series of quality control checks before assembling the reads against a reference human sequence. This is followed by a local re-alignment to correct alignment errors due to insertions/deletions (indels) and re-calibration of quality scores. The data are now ready for annotation of SNPs and indels as well as interpretation of their effects. This is a comprehensive and thorough pipeline, which makes use of a series of FLOSS utilities.
![]() FastQC [18] – FastQC aims to provide a simple way to do some quality control checks on raw sequence data coming from high-throughput sequencing pipelines. It provides a modular set of analyses, which you can use to give a quick impression of whether your data has any problems before doing any further analysis;
FastQC [18] – FastQC aims to provide a simple way to do some quality control checks on raw sequence data coming from high-throughput sequencing pipelines. It provides a modular set of analyses, which you can use to give a quick impression of whether your data has any problems before doing any further analysis;
![]() Burrows-Wheeler Aligner (BWA)[19] – BWA is an efficient program that aligns the relatively short nucleotide sequences against the long reference sequence of the human genome. It implements two algorithms, bwa-short and BWA-SW depending on the length of the query sequence. The former works for lengths shorter than 200 bp and the latter for longer sequences up to around 100 kbp. Both algorithms do gapped alignment. They are usually more accurate and faster on queries with low error rates;
Burrows-Wheeler Aligner (BWA)[19] – BWA is an efficient program that aligns the relatively short nucleotide sequences against the long reference sequence of the human genome. It implements two algorithms, bwa-short and BWA-SW depending on the length of the query sequence. The former works for lengths shorter than 200 bp and the latter for longer sequences up to around 100 kbp. Both algorithms do gapped alignment. They are usually more accurate and faster on queries with low error rates;
![]() SAMtools [20] – SAM (Sequence Alignment/Map) tools are a set of utilities that can manipulate alignments from files in the BAM format, which is the format of the raw data output file of the sequencers. SAMtools exports to the SAM (Sequence Alignment/Map) format, performs sorting, merging and indexing of the sequences and allows rapid retrieval of reads from any region;
SAMtools [20] – SAM (Sequence Alignment/Map) tools are a set of utilities that can manipulate alignments from files in the BAM format, which is the format of the raw data output file of the sequencers. SAMtools exports to the SAM (Sequence Alignment/Map) format, performs sorting, merging and indexing of the sequences and allows rapid retrieval of reads from any region;
![]() Picard [21] – Picard .http://picard.sourceforge.net/. provides Java-based command-line utilities that allow manipulation of SAM and BAM format files. Furthermore there is a Java API (SAM-JDK) for developing new applications able to read in as well as write SAM files
Picard [21] – Picard .http://picard.sourceforge.net/. provides Java-based command-line utilities that allow manipulation of SAM and BAM format files. Furthermore there is a Java API (SAM-JDK) for developing new applications able to read in as well as write SAM files
![]() VCFTools [22] – the Variant Call Format (VCF, http://vcftools.sourceforge.net/) is a specification for storing gene sequence variations and VCFtools is a package designed for working with these VCF format files, for example those generated by the 1000 Genomes Project. VCFtools provides a means for validating, merging, comparing and calculating some basic population genetic statistics;
VCFTools [22] – the Variant Call Format (VCF, http://vcftools.sourceforge.net/) is a specification for storing gene sequence variations and VCFtools is a package designed for working with these VCF format files, for example those generated by the 1000 Genomes Project. VCFtools provides a means for validating, merging, comparing and calculating some basic population genetic statistics;
![]() BGZip [23] – BGZip is a data compression utility that uses the Burrows-Wheeler transform and other techniques to compress archives, sounds and videos with high compression rates;
BGZip [23] – BGZip is a data compression utility that uses the Burrows-Wheeler transform and other techniques to compress archives, sounds and videos with high compression rates;
![]() Tabix [24] – Tabix is a tool that indexes position sorted files in tab-delimited formats such as SAM. It allows fast retrieval of features overlapping specified regions. It is particularly useful for manually examining local genomic features on the command-line and enables genome viewers to support huge data files and remote custom tracks over networks;
Tabix [24] – Tabix is a tool that indexes position sorted files in tab-delimited formats such as SAM. It allows fast retrieval of features overlapping specified regions. It is particularly useful for manually examining local genomic features on the command-line and enables genome viewers to support huge data files and remote custom tracks over networks;
![]() Variant Effect Predictor [25] – this is a utility from Ensembl that provides the facility to predict the functional consequences of variants. Variants can be output as described by Ensembl, NCBI or the Sequence Ontology;
Variant Effect Predictor [25] – this is a utility from Ensembl that provides the facility to predict the functional consequences of variants. Variants can be output as described by Ensembl, NCBI or the Sequence Ontology;
![]() Genome Analysis ToolKit (GATK) [26] – the GATK is a structured software library from the Broad Institute that makes writing efficient analysis tools using next-generation sequencing data very easy. It is also a suite of tools to facilitate working with human medical re-sequencing projects such as The 1000 Genomes and The Cancer Genome Atlas. These tools include tools for depth of coverage analysis, a quality score re-calibration, a SNP and indel calling and a local sequence re-alignment.
Genome Analysis ToolKit (GATK) [26] – the GATK is a structured software library from the Broad Institute that makes writing efficient analysis tools using next-generation sequencing data very easy. It is also a suite of tools to facilitate working with human medical re-sequencing projects such as The 1000 Genomes and The Cancer Genome Atlas. These tools include tools for depth of coverage analysis, a quality score re-calibration, a SNP and indel calling and a local sequence re-alignment.
10.7.7 Development tools
An integrated development environment (IDE) is a software application that provides comprehensive facilities for programmers to develop software. An IDE normally consists of a source code editor, a compiler, a debugger and tools to build the application. At OGT several different IDEs are currently in use; this is partly due to personal preferences as well as variations in functionality.
![]() NetBeans [27] – the NetBeans IDE enables developers to rapidly create web, enterprise, desktop and mobile applications using the Java platform. Additional plug-ins extend NetBeans enabling the development in PHP, Javascript and Ajax, Groovy and Grails, and C/ C++;
NetBeans [27] – the NetBeans IDE enables developers to rapidly create web, enterprise, desktop and mobile applications using the Java platform. Additional plug-ins extend NetBeans enabling the development in PHP, Javascript and Ajax, Groovy and Grails, and C/ C++;
![]() Eclipse [28] – Eclipse was originally created by IBM and, like NetBeans, provides an IDE that allows development in multiple languages. One advantage Eclipse has over other development platforms is its support of a wide variety of plug-ins that allow the same editing environment to be used with multiple languages including Perl and XML;
Eclipse [28] – Eclipse was originally created by IBM and, like NetBeans, provides an IDE that allows development in multiple languages. One advantage Eclipse has over other development platforms is its support of a wide variety of plug-ins that allow the same editing environment to be used with multiple languages including Perl and XML;
![]() R Studio [29] – R Studio is an IDE purely for R. (R is a statistics processing environment, see Chapter 4 by Earll, for example.)
R Studio [29] – R Studio is an IDE purely for R. (R is a statistics processing environment, see Chapter 4 by Earll, for example.)
10.7.8 Web services
Ubiquitous within any IT offering, data must be presented to consumers and web-based systems offer much in terms of flexibility and speed of development. Currently, OGT uses packages that will be familiar to many readers of this book, namely:
![]() Apache [30] – the Apache HTTP Server Project is an effort to develop and maintain an open source HTTP server for modern operating systems including UNIX and Windows NT. It ships with most Linux distributions and it is our default web server;
Apache [30] – the Apache HTTP Server Project is an effort to develop and maintain an open source HTTP server for modern operating systems including UNIX and Windows NT. It ships with most Linux distributions and it is our default web server;
![]() Tomcat [31] – like Apache, Tomcat is an open source web server but it is also a servlet container, which enables it to deploy Java Servlet and JavaServer Pages technologies and it is for this that it has been used at OGT;
Tomcat [31] – like Apache, Tomcat is an open source web server but it is also a servlet container, which enables it to deploy Java Servlet and JavaServer Pages technologies and it is for this that it has been used at OGT;
![]() PHP [32] – PHP is a widely used general-purpose scripting language that is especially suited for web development as it can be embedded into HTML. In conjunction with Apache and MySQL, we have used it extensively to develop our internal web services.
PHP [32] – PHP is a widely used general-purpose scripting language that is especially suited for web development as it can be embedded into HTML. In conjunction with Apache and MySQL, we have used it extensively to develop our internal web services.
In addition to the free tools listed above, there are also those that require a commercial licence (but can be free to academic and non-profit groups). As this adds to the cost of implementation, each application is evaluated on a case-by-case basis. If a tool can provide additional ‘must-have’ functionality, the investment can often be justified. Below are some commercial non-FLOSS tools that we have employed:
![]() BLAT [33] – the Blast Like Alignment Tool or BLAT is an alternative to BLAST and is available from Kent Informatics, a spin-out of the University of California at Santa Cruz (UCSC). BLAT is similar to BLAST in that is used to find related sequences of interest. However, it has been optimised for finding better matches rather than distant homologues. Thus, BLAT would be a better choice to use when looking for sequences that are stronger and longer homologues. Unlike BLAST, a BLAT query runs against a database that has been pre-loaded into memory. Coupled with BLAT’s ‘close-relatives’ algorithm, the result is a search tool that runs very rapidly. In many applications where distant sequence relationships are not required, BLAT provides a highly efficient approach to sequence analysis. Unlike BLAST, however, BLAT is not free and commercial users require a licence (although it is free to non-profit organisations);
BLAT [33] – the Blast Like Alignment Tool or BLAT is an alternative to BLAST and is available from Kent Informatics, a spin-out of the University of California at Santa Cruz (UCSC). BLAT is similar to BLAST in that is used to find related sequences of interest. However, it has been optimised for finding better matches rather than distant homologues. Thus, BLAT would be a better choice to use when looking for sequences that are stronger and longer homologues. Unlike BLAST, a BLAT query runs against a database that has been pre-loaded into memory. Coupled with BLAT’s ‘close-relatives’ algorithm, the result is a search tool that runs very rapidly. In many applications where distant sequence relationships are not required, BLAT provides a highly efficient approach to sequence analysis. Unlike BLAST, however, BLAT is not free and commercial users require a licence (although it is free to non-profit organisations);
![]() Feature Extraction – Agilent’s Feature Extraction software reads and processes the raw microarray image files. The software automatically finds and places microarray grids and then outputs a text file containing all the feature information. There is currently no FLOSS alternative;
Feature Extraction – Agilent’s Feature Extraction software reads and processes the raw microarray image files. The software automatically finds and places microarray grids and then outputs a text file containing all the feature information. There is currently no FLOSS alternative;
![]() GenePix [34] – GenePix from Molecular Dynamics is another microarray application. It can drive the scanner as well as feature extract data from the resultant image file;
GenePix [34] – GenePix from Molecular Dynamics is another microarray application. It can drive the scanner as well as feature extract data from the resultant image file;
![]() GeneSpring – Agilent’s GeneSpring is an application to process feature extraction files from microarrays used in gene expression studies. It is easy to use and has useful workflows that make it easy analyse this type of data. A disadvantage is that the workflows hide many parameters that might otherwise be adjusted within the analysis. However, performing similar analyses with R is not straightforward and although some in the computational biology group have such a capability there is still, for the moment, at least, the need to licence GeneSpring;
GeneSpring – Agilent’s GeneSpring is an application to process feature extraction files from microarrays used in gene expression studies. It is easy to use and has useful workflows that make it easy analyse this type of data. A disadvantage is that the workflows hide many parameters that might otherwise be adjusted within the analysis. However, performing similar analyses with R is not straightforward and although some in the computational biology group have such a capability there is still, for the moment, at least, the need to licence GeneSpring;
![]() JBuilder [35] – JBuilder is a commercial IDE for software development with Java. This was used initially until other IDEs which were FLOSS became available;
JBuilder [35] – JBuilder is a commercial IDE for software development with Java. This was used initially until other IDEs which were FLOSS became available;
![]() Pathway Studio – the Ariadne Pathway Studio provides data on pathway analyses allowing, for example, the opportunity to view a gene of interest in its biological context and perhaps draw further insights.
Pathway Studio – the Ariadne Pathway Studio provides data on pathway analyses allowing, for example, the opportunity to view a gene of interest in its biological context and perhaps draw further insights.
Lastly, there have been instances when a suitable FLOSS or paid-for option was not available, requiring new software to be commissioned. An example of this occurred when there was a requirement to add additional functionality to our Java-based CytoSure™ software [37]. A clear user need had been identified to provide a method that could take data from comparative genome hybridisations (CGH) and make a call as to the copy number of the regions being analysed. Thus providing a means to translate noisy microarray genomic data into regions of equal copy number. There was a pre-existing algorithm available that could do this but no Java implementation had been created. At times such as this, the bespoke approach is the best way to proceed as this provides the clear advantage of having something tailored to an exact specification.
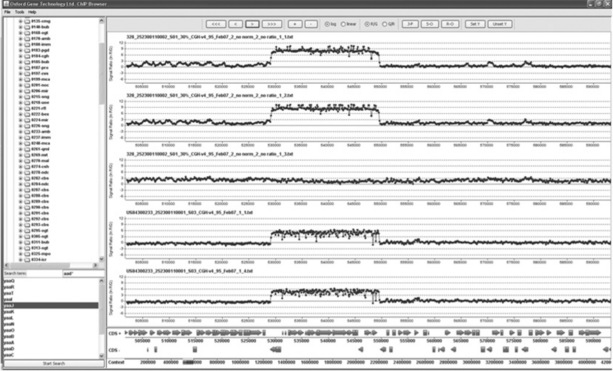
Whereas the tools described above fall into the ‘install and use’ category, there are also many instances when more specialised tools need to be developed. In particular, BioPerl [38] modules for Perl, the equivalent Java library, BioJava [39] provide a firm foundation for development in the bioinformatics area. Similarly, the BioConductor [40] library is invaluable for rapid development of R-based workflows. Although OGT has custom software written in a variety of languages, Perl is perhaps the most common. There are several reasons for this; first, it is one of the easier languages to learn; new colleagues and interns with little prior programming experience can make substantial contributions in a relatively short period of time. Second, the BioPerl set of computational tools is the most mature, often providing much of the core requirements for new scripts. As Perl is predominantly a scripting language, when there is a requirement for a user interface then Java has proved more applicable. Figure 10.4 shows a screenshot from internally developed software designed to view data from ChIP-on-chip microarrays. Our ability to create such software is greatly increased by the availability of free libraries such as BioJava, which was used extensively in this example to process the underlying data.
10.8 Personnel skill sets
The need for bioinformatics programming skills is becoming a necessity within biology and medicine [41]; there is now a requirement for scientists to have access to bioinformatics support and there is currently a lack of such expertise. As reported at the UK parliament by the Science and Technology Committee in their Second Report on Genomic Medicine [42], Professor Sir John Bell stated that ‘the really crucial thing to train in the UK will be bioinformaticians—people who can handle data—the truth is we have now hit the wall in terms of data handling and management’.
This requirement is due to the exponential growth in data being generated, coupled with the increasingly detailed questions being asked of it. Although there are some degree and postgraduate courses in bioinformatics, there is also the opportunity for personal development through self-education and experience. What was once defined as ‘would be nice to have’, bioinformatics programming skills are now more of an ‘essential’ across many fields of biology. This is due to an increased awareness of what may be done and also the massive increase in the amount of data being generated. This all needs to be processed, stored and re-used as required. Similarly, biological information is distributed now thus adding web skills to an already bulging list of requirements in the bioinformatics tool bag.
There are a wide range of skills within the group, and these can be either technical or soft skills. On the technical side there is an absolute requirement to be computer-literate, being comfortable working on the command-line in a Linux environment in the first instance and then building up knowledge and experience with all the built-in tools. Utilities such as the stream editor Sed and the data extraction utility Awk are invaluable in manipulating basic data files but, as per the Perl motto (TIMTOWTDI; pronounced ‘Tim Toady’), there is more than one way to do it. Following on from this is proficiency in at least one programming language; in my experience this is normally a scripting language such as Perl or Python, which as well as being easy to learn have vast repositories of (free) libraries available. The same can be said for other languages such as Java that offer object-orientated desktop and server-based applications. There is a tendency to perhaps ‘forget’ just how rich the landscape of libraries for software developers really is, and how much this has contributed to powerful scientific software.
Of course, as much biological and chemical information is stored in relational databases then knowledge of the Structured Query Language (SQL) is also a key skill for anyone routinely working with such information. Here again, the availability of free database systems (see above) provides the opportunity for anyone to explore and teach themselves without fear of breaking someone else’s database. Free SQL clients such as SQuirrel [33], Oracle SQL Developer [34] and in the MySQL world, MySQL Workbench [35] and PhpMyAdmin [36], provide many capabilities for zero cost. Again, the availability of powerful database management software is incredibly enabling and is another factor aiding companies whose primary goal is science, rather than software development.
Finally, data analysis is also a key skill. More and more, a good understanding of the basics of statistics will help understand the validity and outcome of the many large-scale analyses now available within biology. As such, knowledge of tools, particularly the R project for statistical computing, as well as bioinformatics applications, can be crucial. If more advanced machine-learning approaches are required, the Weka [37] system offers an extensive range of algorithms and techniques, often more than enough to test an initial hypothesis.
10.9 Conclusion
FLOSS fulfils many critical roles within OGT that would otherwise require investment in alternative commercial products. In selecting FLOSS over these other solutions, there is, of course, a cost element to be considered, but that is not the whole story. If any FLOSS product did not meet the required functional criteria then it would not be used; similarly, if a better commercial product becomes available then this would be considered. However, besides reducing expenditure on licences, there are additional advantages from taking a FLOSS approach.
First, if there is a problem, then the very nature of open source software means that, assuming you have the technical ability, it is possible to see directly where the problem is. This was experienced directly at OGT when the format of some NCBI Genbank records changed. The downstream effect of this was that an application using a BioJava file parser threw an error. Because the code was available it was possible to find the problem and fix it locally so the parsing could complete. But beyond that the fix could be passed to the developers for incorporation into the distribution and made available to the whole community. In the event of a problem that we are not able to resolve ourselves then there are usually many other users who can offer a solution or make suggestions on how to proceed. Encountering an error with a commercial product typically requires registration of the error and waiting for someone to do something about it. Even then you may have to wait for the next release before the fix is available to implement.
Similarly, by having access to the source code, there is the opportunity to adapt or extend the functionality to individual requirements as they may arise. In contrast, the functionality of a commercial product is decided by the manufacturers and any additions to this are decided by them alone. FLOSS also avoids the vagaries of a commercial manufacturer, for instance they may choose to amalgamate distinct products into a single offering. This then forces the users to use this new product if they want to keep using the most up-to-date version of the software. We have had experience of this with one piece of software that was incorporated into a more enterprising solution. Interestingly, after a couple of intermediate releases, the standalone product is now being made available. Similarly, commercial vendors control support for older versions of their products and decide when they will stop supporting older releases. Users are powerless to prevent this and helpless to its effects, which can mean they are forced to upgrade even when they would not choose to do so.
In conclusion, FLOSS has significant market share, and performs as well as commercial products with increased flexibility and adaptability. As such, I foresee it retaining a central role in OGTs ’omics platform.
10.10 Acknowledgements
I would like to thank Volker Brenner and Daniel Swan at OGT for reviewing and their helpful suggestions. Also, I would like to thank Lee Harland for his valuable editorial suggestions on the submitted manuscript.
10.11 References
[1] ‘Microarrays: Chipping away at the mysteries of science and medicine’. [Online]. Available: www.ncbi.nlm.nih.gov/About/primer/microarrays.html.
[2] ‘Samba – Opening Windows to a wider world’. [Online]. Available: www.samba.org.
[3] ‘DokuWiki’. [Online]. Available: www.dokuwiki.org.
[4] ‘NetVault – Simplified Data Protection for Physical, Virtual, and Application Environments’. [Online]. Available: www.bakbone.com.
[5] ‘Bacula® – The Open Source Network Backup Solution’. [Online]. Available: www.bacula.org.
[6] ‘Amanda Network Backup’. [Online]. Available: www.amanda.org.
[7] ‘Wikipedia’. [Online]. Available: www.wikipedia.org.
[8] ‘Apache™ Subversion®’. [Online]. Available: http://subversion.apache. org/.
[9] ‘Ensembl’. [Online]. Available: www.ensembl.org/index.html.
[10] ‘National Center for Biotechnology Information’. [Online]. Available: www.ncbi.nlm.nih.gov.
[11] ‘MSPcrunch’. [Online]. Available: www.sonnhammer.sbc.su.se/MSPcrunch. html.
[12] ‘EMBOSS – The European Molecular Biology Open Software Suite’. [Online]. Available: www.emboss.sourceforge.net.
[13] ‘MySQL’. [Online]. Available: www.mysql.com.
[14] ‘PostgreSQL’. [Online]. Available: www.postgresql.org.
[15] ‘Firebird’. [Online]. Available: www.firebirdsql.org.
[16] ‘Oracle’. [Online]. Available: www.oracle.com.
[17] ‘SQL Server’. [Online]. Available: www.microsoft.com/sqlserver.
[18] ‘FastQC – A quality control tool for high throughput sequence data’. [Online]. Available: www.bioinformatics.bbsrc.ac.uk/projects/fastqc/.
[19] ‘Burrows-Wheeler Aligner’. [Online]. Available: http://bio-bwa.sourceforge. net/.
[20] ‘SAMtools’. [Online]. Available: http://samtools.sourceforge.net/.
[21] ‘Picard’. [Online]. Available: http://picard.sourceforge.net/.
[22] ‘VCFtools’. [Online]. Available: http://vcftools.sourceforge.net/.
[23] ‘BGzip-Block compression/decompression utility’.
[24] ‘Tabix – Generic indexer for TAB-delimited genome position files’. [Online]. Available: http://samtools.sourceforge.net/tabix.shtml.
[25] ‘VEP’. [Online]. Available: http://www.ensembl.org/info/docs/variation/vep/ index.html.
[26] ‘The Genome Analysis Toolkit’.
[27] ‘NetBeans’. [Online]. Available: www.netbeans.org.
[28] ‘Eclipse’. [Online]. Available: www.eclipse.org.
[29] ‘RStudio’. [Online]. Available: www.rstudio.org.
[30] ‘Apache – HTTP server project’. [Online]. Available: httpd.apache.org/.
[31] ‘Apache Tomcat’. [Online]. Available: www.tomcat.apache.org/.
[32] ‘PHP: Hypertext Preprocessor’. [Online]. Available: www.php.net.
[33] ‘Kent Informatics’. [Online]. Available: www.kentinformatics.com.
[34] ‘GenePix’. [Online]. Available: www.moleculardevices.com/Products/Software/GenePix-Pro.html.
[35] ‘JBuilder’. [Online]. Available: www.embarcadero.com.
[36] ‘Ingenuity’. [Online]. Available: www.moleculardevices.com.
[37] ‘CytoSure’. [Online]. Available: www.ogt.co.uk/products/246_cytosure_interpret_software.
[38] ‘The BioPerl Project’. [Online]. Available: www.bioperl.org.
[39] ‘The BioJava Project’. [Online]. Available: www.biojava.org.
[40] ‘BioConductor – Open source software for bioinformatics’. [Online]. Available: www.bioconductor.org.
[41] Dudley, J.T. A Quick Guide for Developing Effective Bioinformatics Programming Skills. PLoS Computer Biology. 5(12), 2009.
[42] ‘Science and Technology Committee – Second Report Genomic Medicine’. [Online]. Available: www.publications.parliament.uk/pa/ld200809/ldselect/ldsctech/107/10702.htm.