Investigation-Study-Assay, a toolkit for standardizing data capture and sharing
Philippe Rocca-Serra, Eamonn Maguire, Chris Taylor, Dawn Field, Timo Wittenberger, Annapaola Santarsiero, Alejandra Gonzalez-Beltran and Susanna-Assunta Sansone
Abstract:
This chapter introduces the problems experimentalists in all sectors face in utilizing third party data sets given the unhelpful wealth of formats and terminologies and the consequent mountain of technical frameworks needed to achieve data interoperability. We argue on the importance of a complementary set of open standards, the challenges we must overcome and the role the BioSharing effort is set to play. As an example of progress, we present the open source ISA software solution in action during the curation of the InnoMed PredTox data set, along with its growing active developer and user community, including academia and industrial sectors, such as The Novartis Institutes for BioMedical Research and Janssen Research & Development.
The successful integration of heterogeneous data from multiple providers and scientific domains is already a major challenge within industry. This issue is exacerbated by the absence of agreed standards that unambiguously identify the entities, processes and observations within experimental results.
Empowering Industrial Research With Shared Biomedical Vocabularies, Drug Discovery Today (2011)
7.1 The growing need for content curation in industry
High-quality public bioscience research data should be readily available for re-use in private sector research and development. At present, public resources, diverse in implementation, provide data whose formatting and annotation vary widely, requiring extensive manipulation to open their content to integrative analyses. Such hindrances have motivated community standardization initiatives to develop minimum information checklists, terminologies and file formats, which are increasingly used in the structuring, description, formatting and curation of data sets, although in most cases only within the originating community. These standards aim to ensure that descriptions of entities of interest (e.g. genes, receptors) and related assays contain sufficient contextual information ('experimental meta-data' – e.g. provenance of study materials, technology and measurement types, sample-to-data relationships) to be comprehensible and (in principle) reproducible; without such context, data are of little value.
The process of utilizing shared, publicly available data using community-sourced standards can still test the resolve of even the most ardent advocate [1]. The focus of most community standardization efforts on their own interests or technologies has led to development of equivalent, yet (largely arbitrarily) different localized standards and esoteric repositories, hindering data integration. Whether searching for the scattered files from the various assays in a broadly based study, or assembling the available data on a species or feature of interest, fragmented data sets can only be re-assembled by those equipped to navigate the various terminologies and formats used to represent and annotate their parts (assuming their annotation is sufficient even to reliably identify them), impacting the ability of the R&D community to utilize such data. And, of course, the dearth of accepted standards extends to commercial knowledge providers, whose information also comes in many forms, magnifying the challenge.
Although several integration workflows are routinely run internally, the person-hours cost of 'deciphering' the heterogeneous experimental meta-data still remains significant. Companies must invest significant effort to integrate public bioscience data with their own data; or outsource such activity [2]. The mountain of technical frameworks needed to achieve interoperability between community standards has also hindered the development of general tools. The diversity of standards – and the consequent lack of general tools – hinders discovery, because only a very willing few can even discover, never mind integrate, information scattered across several standalone resources.
7.2 The BioSharing initiative: cooperating standards needed
Left unresolved, or separately and therefore inefficiently addressed by individual companies, the lack of agreed standards will continue to limit the utility of public data. The solution lies in an open collaborative approach between the public and private sectors, lowering individual risk and costs [3, 4]. To establish the lay of the standards landscape, and to build graphs of relationships and complementarities in scope and functionality, the BioSharing community catalogues available standards [5], extending the work started with the Minimum Information for Biological and Biomedical Investigations (MIBBI) portal [6]. These standards often exhibit different levels of maturity and inevitably duplication of effort. Lack of overall coordination also ensures that significant gaps in coverage remain. Although individual communities cannot be corralled into collaboration, the BioSharing initiative is intended to promote those that already exist, discouraging redundant if unintentional competition. In time and after consultation, a set of criteria for assessing the usability and popularity of the standards listed will be implemented, along with links to tools that use them, or data resources annotated with them.
If a common or at least complementary set of standards existed and was widely used by the academic and commercial sectors, routine tasks in the exploitation of both public and proprietary data such as text mining, re-annotation and integration would be greatly facilitated [7]. There are also other benefits accruing to the development and acceptance of general data and reporting standards. For example, by limiting the range and variability of standards, the development and maintenance costs of commercial and academic software come down, resulting in more appropriate resources for the public and private scientific community. In turn, this makes the job of capturing, annotating, integrating, sharing and exploiting data simpler, increasing the prima facie value of the data to others (secondary users) and increasing the return on the investment that supported their generation.
Many challenges lie ahead. Unequivocal 'rules of engagement' must be defined, extensive community liaison managed and rewards and incentives identified for all contributors, whether from the commercial or public sector. In particular, ownership of standards can be problematic in broad collaborations. The appropriate legal framework is still in embryonic form, yet IP rights and licenses must be established to (1) define the boundaries for commercial exploitation by creators and contributors; (2) enable commercial entities to freely contribute time, use cases and requirements; and (3) manage revenues arising from the commercial exploitation of the IP inherent in a resource.
Another critical issue that remains to be addressed is the development of a strategy for the long-term sustainability of this endeavor. Industry funding cannot be the sole source of support, as budgets and priorities fluctuate year to year. Robust relationships among appropriate commercial stakeholders, coupled with participation by governments and research funding organizations will make for a more diversified funding portfolio, buffering the project against fluctuations in the ability of any one partner to contribute. Overall, the cost of implementing this vision is significant and requires BioSharing to continue enlarging its community; particularly, in close partnership with the industry-driven Pistoia Alliance [4].
7.3 The ISA framework – principles for progress
1. Standards initiatives should be more like rafts than cruise liners: simple and unsinkable.
2. As with any evolutionary change, each step should bring reward, not depend on belief.
3. Existing initiatives should be leveraged where possible, for efficiency and buy-in.
4. Solutions should be forward-looking with respect to further, deeper integration.
In combination, these principles require that any solution be simple, immediately beneficial, and respectful of existing work. The 'Investigation/Study/Assay' (ISA) framework, the product of an ongoing collaboration between various research and service groups actively involved in the development of community standards [8], offers such a solution. By providing a generic backbone for structured descriptions of bioscience research – the Investigation, Study, Assay hierarchy around which all else is built – the ISA framework simultaneously offers an interim solution that respects existing data formats and a workable scaffold around which to build new, integrated standards. The basic ISA backbone, extended in an appropriately generic manner, has been implemented as 'ISA-Tab' – a simple format supported by several projects, not least the ISA software suite whose component modules constitute the core elements of a collaborative framework [9].
Using the shared, meta-data-focused ISA framework it is now possible to (1) aggregate investigations of biological systems – where source material has been subject to several kinds of analyses (e.g. genomic sequencing, protein-protein interaction assays and the measurement of metabolite concentrations) – as coherent units of research; (2) perform meta-analyses; and (3) more straightforwardly submit to public repositories, where required. The latter, unfortunately, are still designed for specific analyses types, necessitating the fragmentation of data sets because of the diversity of reporting standards with which the parts must be formally represented.
7.3 The ISA-Tab format
ISA-Tab is the result of a painstaking exercise to map a number of repository submission formats onto one structure for experimental metadata, to facilitate bidirectional conversion; leveraging common elements while intentionally keeping data files external in their native or community formats to side-step interoperability issues. ISA-Tab, illustrated in Figure 7.1, has become parent to a variety of spreadsheet-based formats for data sharing [10]. The Investigation file contains all the information needed to understand the overall goals and means used in an experiment; experimental steps (or sequences of events) are described in the Study and in the Assay file(s). For each Investigation file there may be one or more Study file; for each Study file there may be one or more Assay files.
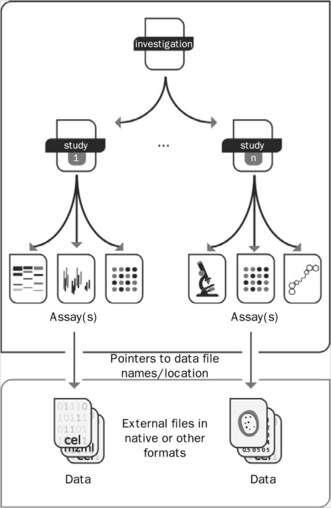
Figure 7.1 Overview of the ISA-Tab format, a general purpose framework with which to collect and communicate complex meta-data (i.e. sample characteristics, technologies used, type of measurements made) from experiments employing a combination of technologies
The Investigation file is intended to meet three needs: (1) to define key entities, such as factors, protocols, parameters, which may be referenced in the other files; (2) to relate Assay files to Study files; and, optionally, (3) to relate each Study file to an Investigation (this only becomes necessary when two or more Study files need to be grouped). The declarative sections cover general information such as contacts, protocols and equipment, and also – where applicable – the description of terminologies and other annotation resources that were used. The Study file contains contextualizing information for one or more assays, for example the subjects studied; their source(s); the sampling methodology; their characteristics; and any treatments or manipulations performed to prepare the specimens. Note that 'subject' as used above could to refer inter alia to an organism, or tissue, or an environmental sample. The Assay file represents a portion of the experimental graph (i.e. one part of the overall structure of the workflow); each Assay file must contain assays of the same type, defined by the type of measurement (i.e. gene expression) and the technology employed (i.e. DNA microarray). Assay-related information includes protocols, additional information relating to the execution of those protocols and references to data files (whether raw or derived).
For example, in a study looking at the effect of a compound inducing liver damage in rats by characterizing the metabolic profile of urine (by NMR spectroscopy) and measuring protein and gene expression in the liver (by mass spectrometry and DNA microarrays, respectively), there will be one Study file and three Assay files, in addition to the Investigation file. The Study file will contain information on the rats (the subjects studied) their source(s) and characteristics, the description of their treatment with the compound and the steps undertaken to take urine and liver (samples) from the treated rats. The Assay file for the urine metabolic profile (measurement) by NMR spectroscopy (technology) will contain the (stepwise) description of the methods by which the urine was processed for the assay, subsequent steps and protocols, and the link to the resultant raw and derived data files. The Assay file for the gene expression profile (measurement) by DNA microarray (technology) will contain the (stepwise) description of how the RNA extract was prepared from the liver (or a section), how the extract was labeled, how the hybridization was performed and so on, and will also contain the links to the resultant raw and derived data files. The Assay file for the protein expression profile (measurement) by mass spectrometry (technology), will contain the (stepwise) description of how the protein extract was prepared from the liver (or a section), how the extract was labeled, how the hybridization was performed and so on, and will also contain the links to the resultant raw and derived data files.
7.3.2 The ISA software suite
The modular ISA software suite, which implements the ISA-Tab format, offers tools to regularize the local management of experimental meta-data, facilitate curation and support conformance to community-defined reporting standards [9]. The modular nature of the ISA software suite separates conformance to standards from the reporting of experimental details (or meta-data), and provides the ability to convert experimental meta-data into various formats (e.g. for submission to some public databases) or to retain it in local storage. The suite's components are variously intended either for experimentalists or those supporting them. The editor tool (ISAcreator) offers familiar spreadsheet-based data entry and facilitates ontology-based curation at source via the BioPortal [11] and the Ontology Lookup Service [12]. Support for conformance to relevant minimum requirements, and the use of specified terminologies, is configurable across the suite (via the support-person-focused ISAconfigurator tool); it can be rigid or flexible to meet in-house requirements. The BioInvestigation Index is a searchable repository through which experimental meta-data and the associated data files can be managed and shared among the users granted access to them (including publicly). Conversion to and from any of a growing number of acceptable formats is enabled by a further module (the ISAconverter tool) [13–15]. The ISA software suite is available for download from the project web site, including the component's technical documentation and users' guide [16].
New collaborative activities continue to move the ISA community along the path to knowledge discovery through broad-scope data integration. For example, work is in progress to (1) augment the ISA code base with Application Programming Interfaces (APIs) to support further collaborative development; (2) facilitate visualization, manipulation and analysis data analysis, informed by the experimental context (the rISA, a R-package for ISA-Tab formatted files) using existing open source analysis platforms [17, 18]; (3) explore cloud-based resource management systems through the ISA suite being deployed on a Bio-Linux platform [19]; and (4) use semantic web approaches to make existing knowledge available for linking, querying, and reasoning in collaboration with the World Wide Web Consortium (W3C) Semantic Web for Health Care and Life Sciences Interest Group (HCLSIG)'s Scientific Discourse task force [20].
7.3.3 The ISA commons
As the collaboration continues to grow and new groups join in, we are on the path to building the ISA commons [21], a growing, exemplar ecosystem of data curation and sharing solutions built on the ISA framework. Rooted in real case studies, this framework is already used by many communities in domains as diverse as environmental health, stem cell discovery, toxicogenomics, environmental genomics, plant metabolomics and metagenomics while maintaining cross-domain compatibility. For example, the Novartis Institutes for BioMedical Research (NIBR [22]) conducts research aimed at drug discovery and development and is developing an instance of selected ISA software components integrated as part of an extended workflow for a microarray gene expression resource. Janssen Research & Development, LLC (http://www.janssenrnd.com) discovers and develops innovative medicines in several therapeutic areas including immunology where the ISA framework is being used to collect, annotate, and search relevant data sets. The use of these software components is aimed at enhancing curation efforts for data integration and analysis of in-house and public data sets. Furthermore, the endorsement of the ISA software by public systems, continued community engagement and growing list of project contributors have engendered a bioscience 'commons' of interoperable tools and data sets [21, 23]. The collaborative ISA framework offers a novel approach to the unsettled status quo by restricting itself to the harmonization of the structure of experimental meta-data only, allowing users of (parts of) data sets to 'connect the metadata dots'. Harmonization of experimental meta-data is important for many resource providers, as our case studies illustrate, for more efficient and better-informed comparison of studies across assays and domains.
To better understand the use of the ISA framework and related developments, we offer the example of the curation process for the European PredTox data sets – a collaborative, distributed study where source material is subject to several kinds of assay in parallel in the search for markers to predict the toxicity of drug candidates [24–27]. The ISA framework was used to enable standards-compliant harmonization and curation prior to the release of study to the public in 2011.
7.3.4 The InnoMed PredTox case study
PredTox [28] was part of the InnoMed Integrated Project, coordinated by the European Federation of Pharmaceutical Industries and Associations (EFPIA [29]), a body representing the research-based pharmaceutical industries and biotech SMEs operating in Europe. InnoMed is a precursor to the Innovative Medicines Initiative, IMI; Europe's largest current public-private initiative [30] with 19 partners (14 pharma companies, three universities, two technology providers). The goal of PredTox was to assess the value of combining results from 'omics technologies with the results from more conventional toxicology methods to support more informed decision-making in pre-clinical safety evaluation. An overview of the PredTox data sets is given in Figure 7.2. The depth and breadth of the PredTox data sets define a landmark for data curation, highlighting the difficulties of structuring multi-assays studies according to common standards.
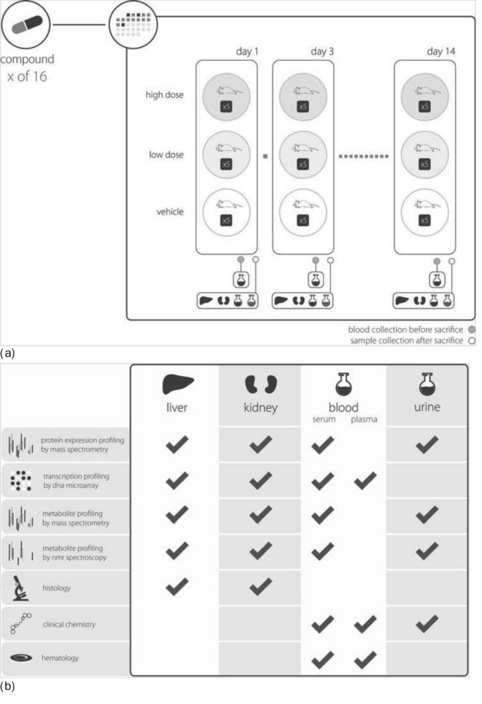
Figure 7.2 An overview of the depth and breadth of the PredTox experimental design (a), and content assay types (b)
Raw and processed data, as well as experimental descriptions, protocols, compounds and sample collection procedures were provided by GeneData [31], a member of the PredTox consortium, in tab-delimited free text, with audit information detailing the person by whom data files were produced or sample processing performed, along with the date. As a first step, the identification schemas devised by the data producers to model animals, samples and aliquots and to link those to data files were disambiguated. Tabular information was then mapped to ISA-Tab syntactic elements, and the data imported by ISAcreator using that mapping, allowing for quick upload.
Sixteen studies were created, one per drug candidate. Experimental descriptions were harmonized to meet the requirement of relevant community-standards, and semantic tagging was performed using OBO foundry ontologies [32] accessed via the ISAcreator ontology widget (Figure 7.3). The Ontology of Biomedical Investigations (OBI [33]) was used for many elements of experimental steps; the vocabulary from the HUPO Proteomics Standards Initiative (PSI) [34] was used to identify and describe mass spectrometers and analytical columns. The same terms were also used for metabolite-focused applications of that analytical technique. Biological annotations relating to organ or tissue were sourced from the Foundational Model of Anatomy (FMA [35]). The drug candidates were submitted to the Chemical entities of biological interest (CHEBI [36]) database and ontology for inclusion. For targeted metabolomics applications, internal standards were used (lactate and trimethylsilyl propionate); those were also tagged using CHEBI as well as all identified metabolites identified and used in the data matrices, whereas units were expressed using Unit ontology [37].
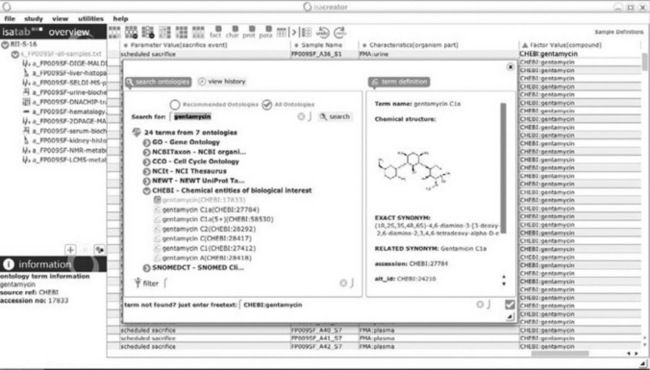
Figure 7.3 The ontology widget illustrates here how CHEBI and other ontologies can be browsed and searched for term selection. Along with the selected term, the internal ontology manager displays the information on each ontology term and records the ID and parent ontology, enabling provenance and tracking
Completing the semantic tagging, raw and processed data files were linked to the detailed description of the studies. Then ISA-Tab formatted files were run through the ISAvalidator, which checked and ensured that all annotation followed the community-defined minimum information requirements set for each of the omics components: MIAME (transcriptomics [38]), MIAPE-MS (proteomics [39]) and CIMR (metabolomics [40]). The completed PredTox data set was then uploaded to a public instance of the BioInvestigation Index hosted at The European Bioinformatics Institute (accession numbers: BII-S-8 to BII-S-23).
7.4 Lessons learned
7.4.1 Open source, collaborative environment
The ISA software solution is a suite of open source and freely available modules, with an active community of users (researchers and curators) and contributing developers. GitHub [41] is the social repository selected to support an environment for code sharing and collaborative development, with the ultimate goal to achieve long-term sustainability and foster self-reliance. Feature requests and bug reports are tracked, discussed and assigned to specific developers.
Due to the modular nature of the ISA software, contributors can take responsibility for the development of specialized modules, contributions to and extensions of the core code. In turn, these are reviewed, vetted or rejected thus making it possible to implement agile practice in an open framework. The main requirement in such open environment is compliance to the open source licensing contract, which is essential to ensure that contributions are duly acknowledged while limiting aggressive open source 'free loading'.
Augmenting the ISA code base with APIs will also support further collaborative development and connections to other widely used data management systems (e.g. [42]). Dealing with feature requests, code forking and subsequent reviews, however, is always a challenging and demanding task, particularly when the ecosystem of users, contributors, and collaborators continues to grow [22, 23].
7.4.2 Curation practices
Generally, even if some good data management practices are in place when data sets are produced, without community standards any subsequent curation or re-use is neither simple nor straightforward. The current crop of minimum information checklists, terminologies and file formats is still growing, lacking control, creating integration headaches especially when new technologies or combinations of technologies are employed. The harmonization of standards remains patchy at best and far from crossing all life science and biomedical domains. Minimum information checklists are often seen as burdensome and over-prescriptive; ontologies too rich and complex; formats intractable. Although the ISA framework can assist in the process of selecting and using the appropriate standards, manual intervention is seldom completely avoidable; our job is to minimize the effort required.
For the IMI InnoMed PredTox, the necessity for well-annotated data and unambiguous meta-data was especially apparent during data analysis. Data sets from the same experimental series were generated in different laboratories, applying diverse technologies, and then had to be integrated for cross-platform and cross-study analyses. Therefore, both the methods and procedures in the laboratories, and the collection and reporting of meta-data had to be highly standardized. The integrated data analysis was performed on three levels. First, profiling data (e.g. transcriptomics) was integrated with conventional data, for example histopathology and serum chemistry, to identify expression markers through phenotypic anchors. Already, this level of integration required reliable identification of animals, treatment regimen, and derived samples that were further processed and used in the different laboratories to yield the said data sets. Second, data from multiple profiling technologies together with conventional data were analyzed across the different tissue types, to identify, for example, how transcriptional changes in the target organ relate to metabolome changes in serum in dependence of histopathological outcome. Third, data from multiple compounds (studies) were integrated to allow for the identification of common mechanisms between compounds causing similar phenotypic (i.e. histopathological) endpoints.
In summary, the IMI InnoMed PredTox generated a rich data set that will be of value to the general public in the evaluations of those highly parallel profiling techniques and in the curation and annotation practice applied using the ISA software suite.
7.5 Acknowledgments
The authors would like to thank all the collaborators who have contributed to the development of the ISA software suite. Special acknowledgement goes to the InnoMed PredTox consortium, for the data sets, and Stephen Marshall, Dorothy Reilly and Stephen Cleaver (of NIBR's Quantitative Biology Unit, Developmental and Molecular Pathways Platform) for the NIBR's case study. The ISA software suite and the BioSharing initiative are currently funded by grants from the Biotechnology and Biological Sciences Research Council and the Natural Environment Research Council's Environmental Bioinformatics Centre.
7.6 References
[1] The Value of Outsourcing Bioinformatics, Bio-IT: http://www.bio-itworld. com/2011/04/21value-outsourcing-bioinformatics.html.
[2] Dealing with data. Science. 2011; 331:692.
[3] Barnes, M., et al. Lowering industry firewalls: pre-competitive informatics initiatives in drug discovery. Nature Reviews Drug Discovery. 2009; 8:701.
[4] Pistoia Alliance: http://www.pistoiaalliance.org/.
[5] BioSharing: http://www.biosharing.org/.
[6] Taylor, C.F., et al. MIBBI: A Minimum Information Checklist Resource. Nature Biotechnology. 2008; 26:889.
[7] Harland, L., et al. Empowering Industrial Research With Shared Biomedical Vocabularies. Drug Discovery Today. 2011; 16:940.
[8] Field, D., et al. 'Omics Data Sharing. Science. 2009; 326:234.
[9] Rocca-Serra, P., et al. ISA software suite: supporting standards-compliant experimental annotation and enabling curation at the community level. Bioinformatics. 2010; 26:2354.
[10] Baker, N., ISA-TAB has become a parent standard for a variety of spreadsheet-based formats for data sharing…' Evaluation of: [Rocca-Serra P et al. ISA software suite: supporting standards-compliant experimental annotation and enabling curation at the community level. Bioinformatics. 2010;26(18) Sep 15:2354–2356, doi: 10.1093/bioinformatics/btq415. [Faculty of 1000, 19 August 2010. F1000.com/4839956].
[11] Noy, N.P., et al. BioPortal: ontologies and integrated data resources at the click of a mouse. Nucleic Acids Research. 2009; 37:W170–W173.
[12] Côté, R., et al. The Ontology Lookup Service: bigger and better. Nucleic Acids Research. 2010; 38(Web Server issue):W155–W160.
[13] Parkinson, H., et al. ArrayExpress update-from an archive of functional genomics experiments to the atlas of gene expression. Nucleic Acids Research. 2009; 37:868.
[14] Vizcaíno, J.A., et al. The Proteomics Identifications database: 2010 update. Nucleic Acids Research. 2010; 38:736.
[15] Shumway, M., et al. Archiving next generation sequencing data. Nucleic Acids Research. 2010; 38:870.
[16] ISA software suite: http://isa-tools.org.
[17] BioConductor: http://bioconductor.org.
[18] Galaxy: http://galaxy.psu.edu.
[19] Cloud Bio-Linux: http://cloudbiolinux.com.
[20] W3C HCLSIG, Scientific Discourse task force, Discourse, Data and Experiment sub-task: http://www.w3.org/wiki/HCLSIG/SWANSIOC.
[21] NIBR: http://www.nibr.com/.
[22] Sansone SA et al. Towards interoperable bioscience data. Nat Genet (accepted).
[23] ISA commons: http://isacommons.org.
[24] Ellinger-Ziegelbauer, H., et al. The enhanced value of combining conventional and 'omics' analyses in early assessment of drug-induced hepatobiliary injury. Toxicology and Applied Pharmacology. 2011; 252:97–111.
[25] Boitier, E., et al. A comparative integrated transcript analysis and functional characterization of differential mechanisms for induction of liver hypertrophy in the rat. Toxicology and Applied Pharmacology. 2011; 252:85–96.
[26] Hoffmann, D., et al. Performance of novel kidney biomarkers in preclinical toxicity studies. Toxicological Sciences. 2010; 116:8–22.
[27] Collins, B.C., et al. Use of SELDI MS to discover and identify potential biomarkers of toxicity in InnoMed PredTox: a multi-site, multi-compound study. Proteomics. 2010; 10:1592–1608.
[28] InnoMed PredTox: www.innomed-predtox.com.
[29] EFPIA: http://www.efpia.org.
[30] IMI: http://www.imi.europa.eu.
[31] GeneData: http://www.genedata.com.
[32] Smith, B., et al. The OBO Foundry: coordinated evolution of ontologies to support biomedical data integration. Nature Biotechnology. 2007; 25:1251.
[33] OBI: http://purl.obolibrary.org/obo/obi.
[34] Martens, L., et al. Data standards and controlled vocabularies for proteomics. Methods Molecular Biology. 2008; 484:279–286.
[35] Rosse, C., Mejino, J.L.V. A reference ontology for biomedical informatics: the Foundational Model of Anatomy. Journal of Biomedical Informatics. 2003; 36:478–500.
[36] Degtyarenko, K., et al. ChEBI: a database and ontology for chemical entities of biological interest. Nucleic Acids Research. 2008; 36:D344–D350.
[37] Unit Ontology: http://www.berkeleybop.org/ontologies/owl/UO.
[38] Brazma, A., et al. Minimum information about a microarray experiment (MIAME)-toward standards for microarray data. Nature Genetics. 2001; 29:365–371.
[39] Taylor, C., et al. The minimum information about a proteomics experiment (MIAPE). Nature Biotechnology. 2007; 25(8):887–893.
[40] Sansone, S.A., et al. The metabolomics standards initiative. Nature Biotechnology. 2007; 25(8):846–848.
[41] ISA on GitHub: https://github.com/ISA-tools.
[42] Szalma, S., et al. Effective knowledge management in translational medicine. Journal of Translational Medicine. 2010; 19(8):68.
