Design Tracker: an easy to use and flexible hypothesis tracking system to aid project team working
Abstract:
Design Tracker is a hypothesis tracking system used across all sites and research areas in AstraZeneca by the global chemistry community. It is built on the LAMP (Linux, Apache, MySQL, PHP/Python) software stack, which started as a single server and has now progressed to a six-server cluster running cutting-edge high availability software and hardware. This chapter describes how a local tool was developed into a global production system.
12.1 Overview
Process management systems and software are now used across many industries to capture, integrate and share pertinent data for a given process. Given the increase in the quantity of data available to project teams in the pharmaceutical industry, as well as the drive to improve productivity and reduce timelines, new approaches and solutions are needed to help the project scientists manage, capture, view and share data across the design – make – test – analyse (DMTA) cycle.
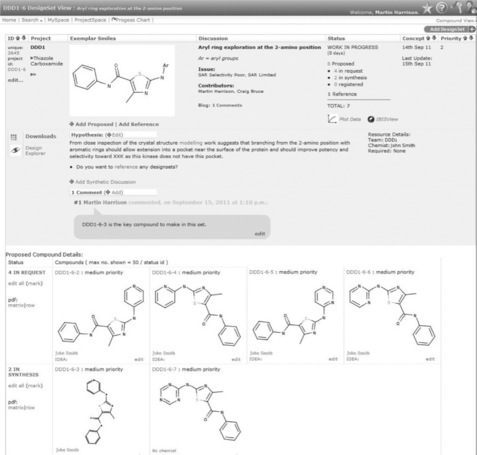
Here, we describe Design Tracker and its software stack that specifically manages some of the various processes that occur in the DMTA cycle [1]. The software allows the tracking of design ideas (hypotheses) and associated compounds from within individual projects or across projects, known as a DesignSet. Candidate compounds associated with a design hypothesis can be prioritised and proposed for synthesis. Synthetic chemists are assigned and it is they who then progress the compounds (and associated design hypothesis) through the complete synthetic work on the design hypothesis. Figure 12.1 shows a sample DesignSet, which includes the hypothesis and proposed compounds; in this case four compounds have been requested and two are currently in synthesis. The compounds are automatically assigned as registered once they appear in the AstraZeneca compound collection and can then be sent for testing. Analysis of the data is then performed by scientists for the specific design hypothesis and the outcome is captured. Based on the outcome and learning, a further round of the DMTA cycle can be started.
Design Tracker stores projects, design hypotheses and compounds in a database together with associated data and creates automated mappings to other AstraZeneca databases to pull in calculated and biological test data for the individual compounds, design hypotheses or projects. Design Tracker creates associations between chemists, compounds, design hypotheses, designers, projects, project issues, project series, etc., to allow different views of the information stored on the database. Views are tailored for individual group requirements and working practices, be they compound designers, synthetic chemists, synthetic team leaders, etc.
Design Tracker is the second generation of software for the capturing of design hypothesis and associated compounds. Design Tracker originated in 2006 when there was a desire for project chemists, medicinal chemists, computational chemists and physical chemists to work together more closely on projects. One of the barriers to this was a lack of clarity about what each discipline was doing. There was a clear need for an online document of the project team’s activities that would allow the project team members to review and input into each other’s work. The original technology chosen was a wiki as it had lots of built-in functionality for templates, revisions, etc. As the individual project wiki pages grew larger, we extracted data from the page and populated a database for easier searching and navigation. This eventually led to the creation of a progress chart for visual planning. This visual planning board concept is still in use today as part of Design Tracker, which is shown in Figure 12.2. The wiki implementation, version 1, was known as the Compound Design Database (CoDD) [2].
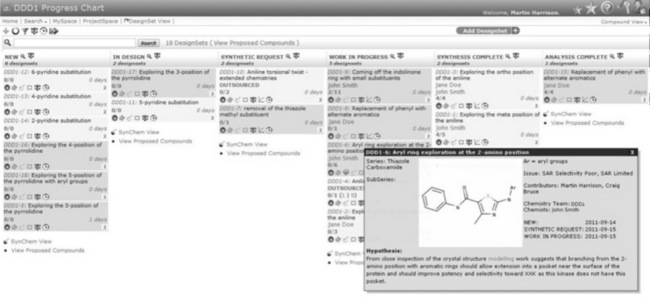
Figure 12.2 The progress chart for the DDD1 project. All DesignSets for the project are visible organised by status. A hover over provides a quick overview for each DesignSet
At the same time within AstraZeneca there was also a move towards ‘hypothesis-based design’. Rather than attempt to enhance design by improving the odds through ever-increasing numbers of compounds, relevant questions were asked of the data and compounds were designed in order to answer these questions. This hypothesis-driven approach allows the prosecution of design systematically, potentially with a reduced number of compounds and design cycles. Clearly, this approach would benefit from the designers from each discipline working more closely together. The wiki-based approach described allowed better communication of design rationale within these teams and was, therefore, being developed and used at just the right time for the business.
The wiki-based approach to capture design hypothesis and associated compounds worked well up to a point but was limited by the fact that the project wiki pages tended to become difficult to manage and navigate. Once the number of hypotheses increased beyond a few tens of design ideas, then the wiki page became large and editing became tricky. It was clear that the concept proved invaluable to projects in capturing their design hypotheses. However, a more bespoke solution was then required to build on this success and allow further information to be added and tracked through the DMTA cycle as well as methods for creating different views into the data.
12.2 Methods
Computational chemistry groups have long had access to Linux environments; it is core to the specialist software used in this field. Compared with the corporate Windows environment, Linux workstations and server systems may be fewer in number. Being dedicated to small R&D groups, there is generally far greater freedom to exploit their full functionality, such as running Apache web servers [3]. This is how we first started to explore web-based applications via Apache on our Linux network. As the technologies behind web sites have developed so have our web sites. Initial HTML hand-coded web sites have been replaced by wikis and content management systems. Although the end result is more functional and Web 2.0-like web sites, they are also far more complicated, utilising databases and multiple frameworks (Javascript, CSS and Ajax, etc.).
Traditionally, all of our applications are thick clients – this works well for our vendors. In-house code tends to primarily consist of scripts or command-line binaries, optimised to run on our high performance computing (HPC) clusters. We do have some GUI applications, but the effort to build a GUI is not trivial. Using web sites offers GUI-like functionality with relative ease. HTML tables are ugly but they can be populated from a database using Perl or Python [4]. New web technologies such as DataTables enable interactive tables [5]. Graphing is another example of improvement. We do not want to write and maintain graph libraries for each language we use. Originally we used Emprise Javascript Charts [6], but have switched from this commercial licence to the open source flot library [7]. The move was partly motivated by functionality we needed already being present in flot. An important aspect for pharmaceutical research is the visualisation of molecules. We are now in the fortunate position to have a multitude of chemistry toolkits to assist. Like the graphing libraries we have no desire to write our own chemistry toolkit. The combination of libraries and toolkits available mean that computational chemists can pick them up and make something to suit our specific needs. Our output is typically intended for our local users or for the computational chemistry community throughout the company, for example a relatively small number. Building web-based applications means every R&D scientist can use our software, without the need for Linux. This has had the unexpected impact of creating a global system, which was originally targeted for a few local users.
12.3 Technical overview
Our initial implementation of CoDD was part of our group wiki, which is a MediaWiki installation [8]. MediaWiki is written in PHP [9] and runs using Apache and MySQL. Our server had everything needed to run this: Red Hat Enterprise Linux [10]. What makes our wiki stand out is the integrated chemistry via the MediaWiki plug-in system. We wrote a small PHP plug-in to enable to the use of < smiles > </smiles > and < az > </az > tags. Both would return an image of the SMILES specified or the AZ number. An AZ number is a reference to a compound in the AstraZeneca compound collection. Figure 12.3 shows the use of the < smiles > tag and how we can alter the size and title of the image.
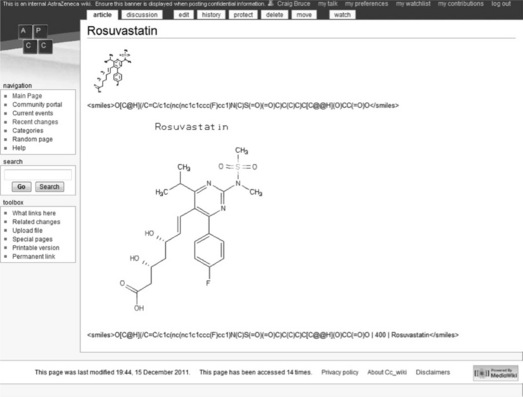
Figure 12.3 Using the smiles tag within our internal wiki. Options such as image dimensions and title can optionally be specified
In this case the PHP MediaWiki plug-in was not doing the chemistry; we actually utilised Python web services. We chose Python because our toolkit of choice, OpenEye, has C++, Java or Python bindings [11]. Python made the most sense in this scenario, as it was straightforward to add to Apache. There are, of course, other toolkits available with different bindings for your language of choice. The web service would either depict the provided SMILES string or perform a MySQL database lookup of the AstraZeneca compound collection to retrieve and depict the relevant SMILES. We wrote various web services – the wiki uses two, Smiles2Gif and AZ2Gif. We have since updated them to write high-resolution PNG images as opposed to the original low resolution GIF images. To achieve this we use Qt, as this gives us additional control over the images, for example the thickness of the bonds, via the pen controller [12].
Once the decision to write Design Tracker was taken we needed to pick the platform to develop on. Given our previous success with Python, we opted to use Django, one of many web frameworks available for Python [13]. Using Python natively meant we could also use the OpenEye toolkits from directly within the application. In terms of the deployment it is the same model as MediaWiki: Apache and MySQL. The only difference is using a Python-Apache link, opposed to PHP-Apache link. Again our single server was able to handle this, but as the popularity of Design Tracker spread across the company’s global sites it became apparent that a more robust solution was required. The existing system had no failover or high availability component; redundant hardware would not protect against MySQL crashing. Our system became locked at fixed and dated versions of all software which was hindering both development and expandability. The decision was taken to invest in a new system to host our numerous Django applications, Design Tracker being the biggest. The platform needed to be robust, scalable, highly available and future-proofed.
At the time Django was a web framework solution that was in its infancy. When we began to start to write Design Tracker, Django version 0.96 was the latest version. What attracted us to Django was the fact that it was open source and was clearly being developed by a relatively large community. There was excellent up-to-date documentation and tutorials online, a roadmap to version 1.0 was clearly defined, and a number of Google groups were discussing Django and offering advice and tips. Most importantly of all, Django is written in Python.
Django also makes the developer create code in a very structured way. This was important as the code base would be written by computational chemists and not dedicated software programmers. Our computational chemists had used python and databases to create web pages before. Moving to Django (or any web framework) was the next step in this evolution to create more advanced web applications.
Django also provides a flexible admin interface that allowed us to quickly investigate different database schemas and relationships. After multiple iterations in this environment, we created the core relationships that still survive today. For pre-existing databases you can use the inspectdb option, as opposed to defining a new schema [14].
Using the admin interface we produced many of the forms and views that enabled us to speak to the customers and stakeholders who would be using Design Tracker. This gave us an opportunity to gather requirements and also to consider capturing additional data that would be useful in further versions. One example of this was the discussions we had with the synthetic chemistry community to try and understand their requirements to support greater usage of the Design Tracker software compared with the CoDD. It was from these efforts and the fact that we could easily capture information using Django’s admin forms that we were able to map out their requirements and how the data would flow through the process. The Django admin views quickly allowed the synthetic chemists and teams to see the potential of developing the software further.
The vision for Design Tracker changed at this point due to this collaboration and we began to develop the software to not only capture design hypotheses and associated compounds but also the ability to assign chemists to compounds and design ideas. The chemists were a part of synthetic teams that were created in Design Tracker and consequently synthetic team views were developed to allow teams to see who was working on what at any given time as well as seeing what compounds and chemistries were coming through the design cycle. This allows synthetic teams and projects to effectively prioritise work based upon resource available and allows designers and synthetic chemists to improve their working relationships through making compound design a more transparent process.
We released version 0.9 (beta release) of Design Tracker at the beginning of September 2008 after four months of development effort, including time to learn Django, by one FTE computational chemist. We started working with a single drug project team to fix bugs for six weeks, releasing minor updates on a weekly basis. At the end of the six weeks a further project was added and we opened the development to enhancement requests from the projects. We released new versions regularly every two weeks to allow the projects to test the new features and give direct feedback. Using this approach the projects felt an integral part of the development and responded well to being the testers of the code. After ten weeks a further two projects were added to Design Tracker, resulting in four projects entering data and tracking their design hypotheses and compounds through the DMTA cycle. One of these projects was being run at another site, which allowed us to gain feedback and enhancement requests from a remote location before we released version 1.0 across multiple sites. In the two months before releasing to the wider community we again released on a two week schedule.
After essentially 16 weeks of beta release testing and enhancements we released version 1.0. This release included documentation and training materials – essentially tutorials for designers and synthetic chemists. Training and support are now carried out by the development team and the super user network [15]. The super user network is sponsored by the PCN (Predictive Chemistry Network) and is represented on each R&D site, unlike the development team. At this stage we did not formally announce the release but allowed Design Tracker to grow by word of mouth. We ran formal training sessions for on-site users in designated training rooms. This involved users going through a series of scenarios on a training version of Design Tracker so users could add, delete and update without fear of corrupting any production data.
Other R&D sites began to request access more regularly so a similar approach was adopted to start with a single project on that site, give demonstrations of the software to a limited group of people, encourage use by responding to their requests for training and minor enhancements promptly and develop relationships with a few key users on each site. The way the different chemistry disciplines were starting to interact with each other gave us the idea to develop the collaborative aspects of the software including commenting (akin to blogs) and voting systems. Design Tracker was starting to become a social network for collaborative drug design.
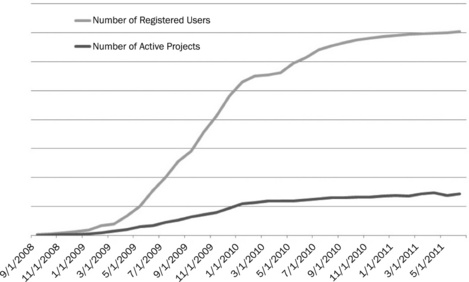
Throughout 2009 we released regular updates to the software and by the end of 2009 there were a large number of projects and registered users, as shown in Figure 12.4. At this stage at some research areas and sites all projects were using Design Tracker. It was at this point that a standard operating procedure was agreed by both the designers, synthetic chemists and project teams to give a clearer understanding of each other’s roles and responsibilities. An example of this is that the designers have agreed not to add more compounds once they have moved a DesignSet into Synthetic Request without speaking directly to the synthetic chemists first. This allows chemists to order reagents and develop the route without having to repeat the process when more compounds were added. The synthetic team leaders and chemists can, therefore, have an understanding of the relative worth of each designset and assign resource appropriately.
12.4 Infrastructure
Now we discuss the infrastructure required to deliver this. Our production server is a single server, which cannot provide a robust solution, it was clear we needed multiple servers. In addition other computing design principles became apparent. For example the database should run on different hardware to the application layer. Within the database layer read and write operations should be restricted to distinct MySQL instances to improve performance. In the application layer dynamic and static requests should be separated. Additionally hardware should be redundant. Software must manage high-availability, loads, failover and all seamlessly work together. This is not new: Wikipedia, Facebook and Google all have similar infrastructures to deliver the most accessed web sites in the world using a similar software stack to us.
Our initial server was a RAID1 system disk (RAID1 requires two disks so the complete data is mirrored at all times), dual power supplies, dual fibre cards, dual network ports, fibre access to the SAN for MySQL binlogs and backups. MySQL binlogs store every SQL statement run since the last backup and can be used to restore a corrupted database in conjunction with the last SQL backup.
Our new cluster consists of blades and storage in one chassis. Shared storage is provided via SAS (serial attached SCSI) connections to each blade. As well as RAID5 a parallel file system is used: GPFS, IBM’s General Purpose file system [16]. The chassis includes dual power supplies, dual network switches and dual SAS switches. Ultimately, the hardware and system software choices are not important, provided that they enable system redundancy and remove any single point of failure. Our choices are based on previous experience and expertise held by our group. Although we want the same basic software stack – Linux, MySQL, Apache, Python – we need to expand it to handle the complexities of failover. One key design decision was to split the database and application operations. Now we have three blades running Apache to handle the application layer and three blades for the MySQL database.
There are multiple topologies available to design a MySQL cluster. We opted for master–slave, in our case a single master and two slaves. Additional software is required to handle this cluster. After evaluating several solutions we opted for Tungsten Enterprise from Continuent [17]. Tungsten Replicator sits on our MySQL blades and ensures the transactions carried out on the master are also repeated on the slaves. Tungsten Connector sits on the application blades and is where all the MySQL connections point to. Depending on the request (read or write) it then directs to the master for writing and a slave for reading. Separating the read and write commands offers substantial performance benefits. As the system grows, further slaves can be added to accommodate more traffic. If the master crashes one of the slaves is promoted automatically. The ability to move the master around means zero down-time for upgrades and maintenance.
For the application layer the setup is more complex than the database layer. We need highly available web servers that can scale. We have long used Apache and continue to do so. Using mod_wsgi we can link to Python [18]. Our application blades are setup with Apache but there is no failover. Nginx is an upcoming web server, which offers a proxy function [19]. We run a single instance of Nginx, which is the point for all URL requests. It then splits requests between the Apache servers. To improve the performance of the web server, dynamic and static requests should be split. Django makes many dynamic requests, and we let Apache handle these. However, Nginx is excellent at serving static files up so it now handles these requests. In this model we can add more Apache servers later as required, and again it allows easy maintenance.
Both Nginx and Tungsten Connector run as a single instance, there is no high availability protection. To ensure these services are always running we use Red Hat Cluster Suite [20]. This ensures that if the blade crashes, they are restarted on another blade. By abstracting Apache under Nginx, we are free to alter web servers later. The open source world changes far more quickly than enterprise software so having the flexibility and freedom to switch the building blocks of our system is paramount. We anticipate that several software components may change within the next few years. Importantly, unlike our previous system we can add more hardware as required and switch software as required. A large upgrade and migration cannot be imposed by external factors.
Although we have opted to use MySQL, OpenEye and Python, it is easy to mix databases, toolkits or languages of choice. Applications like Nginx, Apache and Red Hat Cluster Suite are not tied to a single software stack.
12.5 Review
The next steps for Design Tracker are introducing memory caching via memcached in the application layer for faster performance. MySQL 5.5 is beginning to utilise memcached as well. Currently we use MySQL 5.1 but Tungsten will let us run multiple versions while we upgrade the cluster at our own pace. We are keen to upgrade to the latest version, but with a global production system we test thoroughly on the development cluster first.
Looking forward we will investigate master-master replication across our geographical sites for disaster recovery purposes and to provide enhanced database performance where the limiting step is a slow network connection between sites. We are not tied to MySQL – we could switch to PostgreSQL – and Continuent is building support for other databases as well, including Oracle. From the application perspective would should add more Nginx and Tungsten Connector instances and use the LVS (Linux Virtual Server [21]) to load balance. This has the drawback of requiring more servers to setup LVS, therefore another solution which we are also investigating is using a hardware load balancer which would host the virtual IP addresses then distribute requests to the multiple application servers, each running their own copy of Ngnix and Tungsten Connector.
Design Tracker as an application is widely used and is now a key component of our DMTA cycle. We work constantly to tighten integration of it with other in-house systems. Our users provide a constant stream of enhancement requests, which serve to make Design Tracker better. As we progress we now have a platform that can accommodate our current and future needs.
12.6 Acknowledgements
Design Tracker has been made possible by the efforts and funding of the computational chemistry groups at Alderley Park with notable contributions from Richard Hall, Huw Jones, Graeme Robb, David Buttar, Sandra McLaughlin, David Cosgrove, Andrew Grant and Oliver Böcking.
12.7 References
[1] Plowright, A.T., Johnstone, C., Kihlberg, J., Pettersson, J., Robb, G., Thompson, R.A. Hypothesis driven drug design: improving quality and effectiveness of the design-make-test-analyse cycle. Drug Discovery Today. 2012; 1–2:56–62.
[2] Robb, G., Hypothesis-driven Drug Design using Wiki-based Collaborative Tools Paper presented at:. UK QSAR & Chemoinformatics Spring Meeting, Sandwich, Kent, UK, 2009.
[3] Apache Software Foundation, Apache HTTPD Server Project, November 29, 2011. Available at: http://httpd.apache.org [Accessed 29 November 2011].
[4] Python Software Foundation, Python, November 29, 2011. Available at: http://www.python.org [Accessed November 2011].
[5] Jardine, A., DataTables, November 29, 2011. Available at: http://datatables.net [Accessed November 2011].
[6] Emprise Corporation. Emprise JavaScript Charts. Available at: http://www.ejschart.com. Accessed 29 November 2011.
[7] Laursen O. Google Code: flot. Available at: http://code.google.com/p/flot. Accessed 29 November 2011.
[8] Wikimedia Foundation. MediaWiki. Available at: http://www.mediawiki.org/wiki/MediaWiki. Accessed 29 November 2011.
[9] The PHP Group. PHP. Available at: http://www.php.net. Accessed 29 November 2011.
[10] Red Hat, Inc. Red Hat Enterprise Linux. Available at: http://www.redhat.com/rhel. Accessed 29 November 2011.
[11] OpenEye Scientific Software, Inc. OpenEye. Available at: http://www.eyesopen.com. Accessed 29 November 2011.
[12] Nokia Corporation. Qt. Available at: http://qt.nokia.com. Accessed 29 November 2011.
[13] Django Software Foundation. Django project. Available at: https://www.djangoproject.com. Accessed 29 November 2011.
[14] Django Software Foundation. Django Documentation. Available at: https://docs.djangoproject.com/en/1.3/ref/django-admin/#django-admin-inspectdb. Accessed 29 November 2011.
[15] Cumming J, Winter J, Poirrette A. Better compounds faster: the development and exploitation of a desktop predictive chemistry toolkit. Drug Discovery Today; in press.
[16] International Business Machines Corporation. I BM General Parallel File System. Available at: www.ibm.com/system/software/gpfs. Accessed 29 November 2011.
[17] Continuent Inc. Continuent. Available at: http://www.continuent.com. Accessed 29 November 2011.
[18] Dumpleton G. Google Code: mod_wsgi. Available at: http://code.google.com/p/modwsgi. Accessed 29 November 2011.
[19] Sysoev I. Nginx. Available at: http://nginx.org. Accessed 29 November 2011.
[20] Red Hat, Inc. Red Hat Cluster Suite. Available at: http://www.redhat.com/rhel/add-ons/high_availability.html. Accessed 29 November 2011.
[21] Zhang W. Linux Virtual Server project. Available at: http://www.linuxvirtualserver.org. Accessed 29 November 2011.